-
小分子分泌型多肽是重要的胞间信号分子,广泛地参与调节植物生长发育、抗病和抗逆等生理过程[1-2].快速碱化因子(Rapid alkalinization factor,RALF)属于植物小分子多肽的一种,最早是由Pearce等[3]从烟草叶片提取物中发现,该蛋白能引起烟草悬浮细胞的培养基快速碱化,过表达时能抑制番茄和拟南芥根的生长和发育.至今已在51种植物中鉴定出795个RALF基因[4].研究表明烟草NaRALF基因沉默后主根更长且毛状体细胞发育成不正常的根毛[5].蒺藜苜蓿根中过表达MtRALF1基因时结节发育不正常[6].在细胞悬浮培养基中加入纯化的甘蔗SacRALF1蛋白能抑制愈伤组织细胞的延伸,在培养基加入AtRALF1导致黑暗条件下萌发的拟南芥幼苗下胚轴伸长受到抑制,幼苗放到没有AtRALF1的培养基时下胚轴的伸长得到恢复[7].拟南芥中过表达AtRALK8基因可以显著增强植株抗干旱和线虫侵染的能力[8].拟南芥AtRALF34调控中柱鞘形成细胞的分化进而影响侧根发育[9].拟南芥AtS1P切割内源的AtRALF23以抑制植物免疫响应,而AtFER可以促进配体诱导的AtEFR,AtFLS2和AtBAK1形成复合体进而激活免疫信号[10].
在生殖发育方面,人工合成的番茄SlPRALF蛋白能抑制花粉管的伸长,但不影响花粉水化和花粉活力[11].拟南芥AtRALF4可强烈抑制花粉萌发[12].拟南芥花粉AtRALF4/19与BUPS1/2和ANX1/2形成复合体维持花粉管的完整性,柱头AtRALF34与AtRALF4/19竞争性结合BUPS-ANX复合体,在纳摩尔水平就可以引起花粉管破裂[13]. Shi等[14]从白菜中鉴定出38个RALF基因,有14个在核雄性不育和可育性材料间差异表达.前期笔者通过甘蓝转录组测序从自交不亲和甘蓝“A1”花粉中获取了一个RALF基因的编码序列,命名为BoRALFA1,在甘蓝02-12系基因组CDS数据库(http://brassicadb.org/brad/)中检索不到同源序列.本文从甘蓝“A1”克隆了BoRALFA1的CDS序列,进行了生物信息学分析,对BoRALFA1及其同源基因组织表达模式进行了分析,以期为深入研究的BoRALFA1基因的生物学功能提供基础.
HTML
-
甘蓝高度自交不亲和系“A1”种植在西南大学十字花科歇马实验基地隔离网内. 2019年3月底甘蓝花期,采集开花当天且长势一致的柱头、花粉、萼片、花瓣和叶片,置于-80 ℃冰箱保存备用.
利用RNAprep Pure Plant kit(天根,北京)提取RNA,经琼脂糖电泳检测RNA的完整性,用NanoVue Plus超微量分光光度计(Gen,美国)测定RNA的浓度,然后采用Primescript RT Reagent Kit(TaKaRa,日本)进行反转录合成cDNA的第一条链,-80 ℃冰箱保存备用.以叶片为材料,利用Plant Genomic DNA Kit(天根,北京)提取甘蓝基因组DNA,-20 ℃冰箱保存备用.
-
利用前期甘蓝转录组测序获得的BoRALFA1序列在NCBI(http://www.ncbi.nlm.nih.gov/)和BRAD(http://brassicadb.org/brad/)数据库进行BlastN检索,依据检索结果用Primer Primer 5.0软件设计基因特异性引物RALF-1和RALF-2. RALF-1的序列为ATTAATAATAATATGGGGATGTCTGAAAG,RALF-2的序列为TGGTACTCTACTTGGTCGATTCACA.以甘蓝花粉cDNA和基因组DNA为模板,参照PrimerSTAR Max DNA polymerase说明书配制25 μL反应体系. PCR循环参数为:98 ℃预变形3 min;98 ℃变性15 s,55 ℃退火15 s,72 ℃延伸30 s,35个循环;72 ℃延伸5 min. PCR扩增产物经1%琼脂糖电泳,回收目的条带送重庆擎科公司测序.
-
利用在线软件MultAlin(http://multalin.toulouse.inra.fr/multalin/multalin.html)进行cDNA和gDNA序列比对,用DNAstar软件推导BoRALFA1基因编码的氨基酸序列及其理化性质.利用在线软件SOSUI(http://harrier.nagahama-i-bio.ac.jp/sosui/sosui_submit.html)分析蛋白的跨膜运域.利用在线软件SignalP-5.0(http://www.cbs.dtu.dk/services/SignalP/)分析信号肽序列,利用在线Prosite(https://prosite.expasy.org/)分析蛋白活性位点.用BRAD在线数据库进行BlastN检索,分析甘蓝02-12系基因组BoRALFA1基因编码位点,检索白菜、油菜、拟南芥和琴叶拟南芥基因组中高度同源序列.利用在线数据库Phytozome 12(https://phytozome.jgi.doe.gov/pz/portal.html#)查找Brassica oleracea capitata V1.0基因组中同源序列.多序列比对利用MultAlin软件进行,参照高启国等[15]的方法利用MEGA软件并采用邻接法构建系统发育树.利用在线软件SWISS-MODEL(https://swissmodel.expasy.org/),以AtRALF8空间结构为模板,构建甘蓝BoRALFA1d的空间结构,用DeepView软件查看和分析生成的空间结构.
-
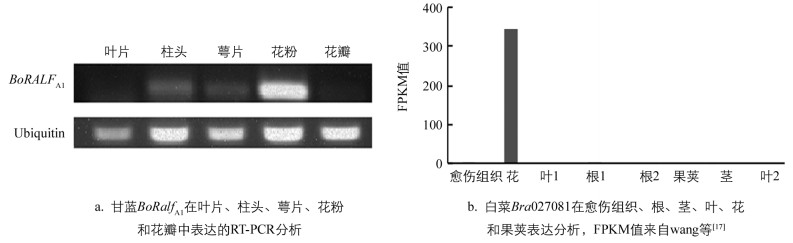
以Ubiquitin作为内参基因,RT-PCR分析甘蓝BoRALFA1基因在柱头、花粉、萼片、花瓣和叶片5个组织的表达情况,PCR反应体系和循环参数参照1.2,其中循环为30个. Ubiquitin基因的循环参照Gao等[16]的方法,循环数为22.基于Wang等[17]对白菜Chiifu-401-42的愈伤组织、根、茎、叶、花和果荚6个组织的转录组分析,从GEO数据库(http://www.ncbi.nlm.nih.gov/geo/)下载转录组分析结果(收录号:GSE43245),利用FPKM值绘制白菜Bra027081基因在6个组织中表达的柱形图.
1.1. 供试材料
1.2. 引物设计和目的基因克隆
1.3. 甘蓝BoRALFA1基因生物信息学分析
1.4. 甘蓝BoRALFA1和白菜Bra027081基因表达分析
-
依据前期转录组测序结果设计特异引物RALF-1和RALF-2,从自交不亲和甘蓝“A1”花粉cDNA和基因组DNA中扩增其cDNA和gDNA序列.测序后序列比对分析表明,BoRALFA1基因没有内含子,其全长CDS为240 bp,共编码79个氨基酸,理论分子量为8.73 kD,等电点为7.80,其中包含9个碱性氨基酸残基(K和R)、8个酸性氨基酸残基(D和E)、25个疏水性氨基酸残基(A,I,L,E,W和N)和23个极性氨基酸残基(N,C,Q,S,T和Y)(图 1).
在线软件SOSUI分析表明,BoRALFA1蛋白包含1个跨膜结构域,其序列为第4位丝氨基酸位到第25位谷氨酰胺.在线软件SignalP-5.0分析表明,BoRALFA1蛋白具有一个N-端信号肽,可信值达0.992 7,最可能的前体蛋白剪切位点为第28位丝氨酸和第29位精氨酸之间.通过在线PROSITE分析发现:BoRALFA1蛋白含有1个N-端豆蔻酰基化位点,位于第2到第7位氨基酸;2个蛋白激酶(PKC)磷酸化位点,分别位于第6到第8位氨基酸以及第51到第53位氨基酸;1个酪蛋白激酶2(CK2)磷酸化位点,位于第24到第27位氨基酸;1个酪氨酸激酶(TYP)磷酸化位点,位于第53到第60位氨基酸.
利用BoRALFA1基因序列在甘蓝02-12系基因组数据库进行BlastN检索,发现该基因定位在C09号染色体上,且在该染色体长度为14 134 bp的片段上存在6个BoRALFA1基因的完整编码框串联排列,分别位于C09:34543886..34544125,C09:34546665..34546904,C09:34549444..34549683,C09:34552223..34552462,C09:34555002..34555241和C09:34557781..34558020,多序列比对表明6个位点编码的序列与甘蓝“A1”中BoRALFA1基因序列完全一致,没有核苷酸差异(图 2).
-
利用甘蓝BoRALFA1基因cDNA序列在芸薹属基因组CDS数据库进行BlastN检索,结果显示甘蓝02-12系基因组中没有注释的相似序列.白菜Chiifu-401-42基因组中Bra027081基因与BoRALFA1的一致性最高,为97%.油菜中有3个基因GSBRNA2T00070572001,GSBRNA2T00020917001和GSBRNA2T00070573001,均与BoRALFA1的一致性较高,分别为99%,98%和97%.在NCBI数据库检索结果表明,BoRALFA1基因cDNA与已报到的油菜RALFbn基因(GeneBank登录号:KC149515)和青花菜BoRALF1基因(GeneBank登录号:DQO59310)完全一致.在拟南芥基因组数据库中检索发现BoRALFA1与AtRALF9和AtRALF15的相似性较高,分别为77%和75%.下载已完成功能分析的拟南芥AtRALF4,AtRALF19和AtRALF34,烟草NaRALF和NtRALF,马铃薯ScRALF3以及杨树PtdRALF1和PtdRALF2的蛋白序列,进行多序列比对,结果见图 3.甘蓝BoRALFA1与油菜RALFbn和GSBRNA2T00070572001、青花菜BoRALF1氨基酸序列完全一致,与上述已进行功能研究的8个RALF蛋白相比,BoRALFA1及其高度同源氨基酸序列的N-端明显较短,且缺少N-保守的RRXL基序[18].在所有的氨基酸序列中C-端序列的相似性较高,有4个高度保守的半胱氨酸、3个高度保守的基序(分别为YIXY,YXRGC以及RCG).
利用BoRALFA1氨基酸序列在琴叶拟南芥基因数据库中进行BlastP检索,结果显示scaffold_200432.1和scaffold_200431.1与BoRALFA1一致性最高,分别为69%和65%.在甘蓝Brassica oleracea capitata V1.0基因组数据库中进行BlastP检索,结果显示Bo2g011740与BoRALFA1蛋白序列相似性最高为58%.将来自甘蓝、白菜、油菜、拟南芥和琴叶拟南芥中与BoRALFA1相似性较高及其上述已进行功能研究的23个RALF蛋白序列构建系统发育树(图 4).结果显示,共形成了2个主要分支,分支A和B.其中甘蓝的BoRALFA1与来自青花菜、白菜和油菜3个物种的5个高度相似性的基因聚合在分支A2中,表明该基因的分化可能早于物种的分化.已完成功能研究来自拟南芥、烟草、杨树和马铃薯的7个基因聚合在分支B中,与BoRALFA1的遗传关系较远.
-
为了进一步确认BoRALFA1基因在甘蓝中的表达情况,分别提取了甘蓝“A1”开花当天柱头、花粉、萼片、花瓣和叶片的总RNA,然后进行RT-PCR分析.产物的凝胶电泳结果表明BoRALFA1基因在甘蓝“A1”花粉中表达量较高,在柱头和花萼中表达量较低,在花瓣中表达量极低,在叶片中几乎没有表达(图 5a).甘蓝BoRALFA1基因与白菜Bra027081基因相似度达97%,系统进化分析表明两者在进化上高度保守. Wang等[17]2011年进行了白菜Chiifu-401-42的愈伤组织、根、茎、叶、花和果荚6个组织的转录组分析,笔者从GEO数据库下载其转录组结果,从中分析Bra027081基因在6个组织表达情况,结果表明Bra027081基因主要在花中表达(图 5b).拟南芥中AtRALF9与BoRALFA1基因相似性最高,笔者利用TRAVA网站分析发现AtRALF9基因也主要是在花粉中表达.
-
利用在线软件GOR分析表明,BoRALFA1蛋白的二级结构由37.97%的α-螺旋(30个氨基酸)、45.57%的无规卷曲(36个氨基酸)和16.46%的延伸主链(13个氨基酸)组成. Frederick等[19]分析了AtRALF8蛋白的三维空间结构. BoRALFA1和AtRALF8氨基酸序列长度仅存在1对氨基酸差异,分别为79和80个氨基酸,两者全长一致性为59%.利用在线软件SWISS-MODEL,以AtRALF8为模板生成BoRALFA1三维结构.其包含了BoRALFA1成熟肽全部序列,空间上类似于大写字母“M”,序列上高度保守的基序YIXY位于N-端起始位置,裸露于外面,YXRGC和RCG基序分别位于靠近C端“M”顶角的内外两侧,N-端YIXY基序是RALF蛋白与受体结合的核心功能域,C-端可以增强N的活力[20-21](图 6).
2.1. 甘蓝BoRALFA1基因序列与编码位点分析
2.2. 甘蓝BoRALFA1同源基因分析
2.3. 甘蓝BoRALFA1和白菜Bra027081基因表达分析
2.4. 甘蓝BoRALFA1蛋白空间结构分析
-
RALF蛋白作为重要的胞间信号因子广泛参与植物的多种生理过程.然而至今关于甘蓝RALF蛋白还鲜有报道.本文从甘蓝“A1”材料中克隆的1个RALF蛋白编码基因,命名为BoRALFA1,其全长CDS为240 bp,编码79个氨基酸,包含1个信号肽和5个翻译后修饰位点.在甘蓝02-12系基因组C09号染色体上存在6个串联重复编码位点.序列比对表明BoRALFA1具有RALF蛋白家族典型保守基序[4],包含4个保守的半胱氨酸残基、N-端YIXY基序、C-端YXRGC和RCG基序.与已经功能鉴定的拟南芥AtRALF4,AtRALF19和AtRALF34[13],烟草NaRALF和NtRALF[3-5],及杨树PtdRALF1和PtdRALF2[22]的氨基酸序列相比,BoRALF1蛋白N-端明显较短,且缺失保守的RRXL基序,该基序是RALF前体蛋白加工过程中丝氨酸蛋白酶AtSIP1的识别位点[18, 23],由此推测BoRALFA1可能存在不同的加工过程.
甘蓝BoRALFA1与白菜Bra027081、油菜RALFbn和青花菜BoRALF1序列高度一致且在进化上高度保守,表明它们可能具有相似的生物学功能.李焰焰等[24]分析表明油菜RALFbn主要在雄蕊中表达,而在雌蕊、花瓣和萼片中没有表达. Zhang等[25]从青花菜中克隆了BoRALF1基因,发现其主要在花粉中表达. Wang等[17]转录组分析显示白菜Bra027081主要在花中表达,Shi等[14]进一步分析表明白菜Bra027081在可育材料花蕾中高量表达,在核雄性不育材料花蕾中不表达.本文结果显示甘蓝BoRALFA1主要在花粉中表达.因此推测BoRALFA1可能在花粉发育和育性方面具有重要的功能. BoRALFA1蛋白具有保守的N-端YIXY基序,在三维空间结构上裸露于表面.研究表明YIXY基序是RALF蛋白与受体结合的核心功能域[20-21].在已经鉴定的与生殖有关的RALF蛋白中,AtRALF4/19与BUPS-ANX受体复合体结合维持花粉管的完整性,AtRALF34与AtRALF4/19竞争性结合BUPS-ANX复合体,致使花粉管破裂[13-26].先前研究显示拟南芥FEROINA (FER)与AtRALF1作用调控主根细胞伸长,与AtxRALF23作用调控植物免疫反应[10-27],最新研究表明AtRALF1-FER调控拟南芥的开花时间[28].因此,接下来工作的重点是进一步明确BoRALFA1蛋白的功能,分离其互作蛋白并探究其作用机理.










 DownLoad:
DownLoad: