-
1997年, Wilmut等[1]通过利用体细胞核移植技术成功地克隆出了世界上第一只体细胞克隆绵羊多利(Doly), 它的诞生证明了高度分化的体细胞仍然具有全能性. 自此以来, 许多哺乳动物物种已经被成功克隆[2]. 目前, 体细胞核移植技术虽然已取得了明显进展, 但还存在克隆效率比较低、克隆动物体型偏大、出生的克隆动物体不易存活等问题.
猪作为一种良好的经济动物, 也是最接近人的模式生物, 其SCNT(Somatic Cell Nuclear Transfer)的低效率和后代异常限制了其在实践中的应用[3]. 这些问题是由许多原因造成的, 目前人们主要的关注点是体细胞重编程不完全对体细胞核移植胚胎发育的影响[4]. 为了提高SCNT的效率, 包括使用不同类型的供体细胞[5]改善去核程序[6], 延长从融合到激活的间隔[7-8]等多种方法被用来调节供体细胞或克隆胚胎的表观遗传修饰.
包含SET和MYND域的蛋白质3(SMYD3)是一种组蛋白H3赖氨酸4甲基转移酶, 在转录调控中起重要作用. 多年来研究者在某些癌细胞系中研究了SMYD3的表达, 在卵母细胞[9]和胚胎中的作用也逐渐被揭示. Bai等[10]发现SMYD3基因能够提高牛成纤维细胞iPSCs诱导效率, 具有体细胞重编程作用. 而在早期胚胎发育中, SMYD3也具有重要的作用[10-12]. 本研究采用RNA干扰(RNAi)和过表达等技术方法, 通过调控德保猪耳成纤维细胞SMYD3基因的表达, 探讨其在猪体细胞核移植过程中的重编程作用, 为提高猪体细胞克隆效率奠定基础.
HTML
-
细胞:德保猪耳缘成纤维细胞来自广西壮族自治区畜牧研究所, 293T细胞和猪颗粒细胞来自于本实验室, 卵母细胞抽取自南宁屠宰场提供的猪卵巢.
质粒:pSicoR-GFP-SMYD3 shRNA来自南京金斯瑞生物科技有限公司, 其他所用质粒均来自本实验室. 猪SMYD3基因shRNA序列为(5′-CTCGAGCCGGCCACAAGCGAGAATGCAAATTCAAGAGATTTGCATTCTCGCTTGTGGTTTTTTGGCGGCCGC-3′).
试剂:X-treme GENE HP DNA Transfection Reagent(Roche公司);Trizol Reagent(Invitrogen公司);DNA聚合酶、qRT-PCR试剂盒(Takara公司);普通质粒提取试剂盒、DNA Markers(天根生化科技有限公司);TransIntreTM EL Transfection(全式金生物技术有限公司). 无特殊说明的其余实验试剂均来源于SIGMA公司.
引物:克隆及定量引物由上海生工公司合成(表 1).
-
在前期实验中我们已分别将pSicoR-GFP-1864和pSicoR-GFP-SMYD3 shRNA表达载体转染细胞, 转染48 h后观察发现空白组细胞无绿色荧光蛋白表达, pSicoR-GFP-1864阴性对照组和pSicoR-GFP-SMYD3 shRNA处理组细胞均有绿色荧光表达. 72 h后收集细胞, 提取总RNA, qRT-PCR结果显示与空白组和阴性对照组相比, SMYD3基因在处理组细胞中表达显著降低(p<0.05), SMYD3基因shRNA干扰效率为54%.
利用转染试剂将本实验室预先构建好并经酶切鉴定的过表达重组质粒pLVX-IRES-ZsGreen1-SMYD3以及干扰重组质粒SMYD3-GFP-shRNA分别与包装质粒NRF、包膜质粒VSVG以5:3:2的比例转染293T细胞, 转染培养72 h后收集病毒.
将猪成纤维细胞以相同的细胞数接种到24孔板中, 当24孔板内的细胞汇合度达到70%时, 向每孔DMEM(Dulbecco's Modified Eagle Medium)培养基里加入病毒以及终质量浓度为8 μg/mL的助感染试剂polybrene. 加入MOI(Multiplicity of Infection)值为5的过表达SMYD3基因病毒细胞作为上调SMYD3组, 加入MOI值为0.8的shRNA病毒作为下调SMYD3组, 未处理的细胞作为空白对照组.
-
用75%酒精对保温取回的卵巢进行消毒, 再用33 ℃生理盐水清洗2~3遍, 用10 mL无菌注射器抽取直径为3~6 mm的卵泡, 在体式显微镜下用捡卵针捡出有3层以上卵丘细胞的卵丘—卵母细胞复合物(Cumulus oocyte complexes, COCs). 把捡出的COCs转移到提前预热的PM(Pericyte Medium)成熟培养盘, 放入38 ℃, 5% CO2, 100%湿度的培养箱中培养.
-
将培养44 h的COCs吸出, 放到提前预热含0.1%透明质酸酶的CCM(Cell Culture Medium)中轻轻吹打, 挑选出含极体的成熟裸卵, 将裸卵放入电融合槽融合和激活, 激活参数为100 V(以每1 mm电激槽宽计)、80 μs、3次电脉冲. 激活后的裸卵移到胚胎培养液PZM-3中, 在38 ℃, 5% CO2, 100%湿度的培养箱中培养, 收集2细胞、4细胞、8细胞、16细胞和囊胚.
-
分别培养未处理、过表达SMYD3基因和抑制SMYD3基因表达的5~7代猪成纤维细胞作为供体. 成熟的卵母细胞如1.3相同方法去除颗粒细胞后, 在镜下采用盲吸法去核. 吸取表面圆滑、形态规则的供体, 然后注入卵母细胞卵周隙内(对于上调和下调SMYD3基因表达的成纤维细胞需在荧光显微镜下挑选绿色荧光蛋白表达的细胞作为供体). 将重构卵进行电激活, 融合/激活参数为83 V(以每1 mm电激槽宽计), 100 μs, 1次直流脉冲. 将融合/激活完的重构卵转移到PZM-3(Porcine Zygote Medium-3)中, 放到38 ℃, 5% CO2, 100%湿度的培养箱中进行培养. 以不同供体进行的核移植, 分为对照组、过表达SMYD3组、抑制SMYD3组.
-
按试剂盒说明提取各组细胞RNA后, 在PCR仪中进行反转录, 反转录条件为25 ℃ 10 min, 42 ℃ 90 min, 95 ℃ 10 min. 以微量反转的cDNA为模板进行qRT-PCR试验, 定量引物序列见表 1, 反应体系见表 2, 反应条件参照说明书. 内参基因是18S, 依据2-ΔΔCT方法计算SMYD3, Nanog和Oct4基因的表达情况.
1.1. 材料
1.2. 细胞转染
1.3. 猪卵母细胞体外成熟
1.4. 卵母细胞孤雌激活
1.5. 核移植
1.6. 微量反转录和qRT-PCR
-
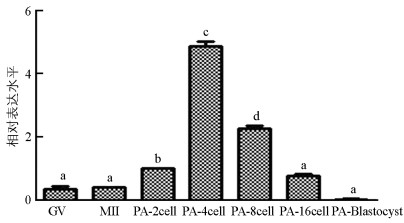
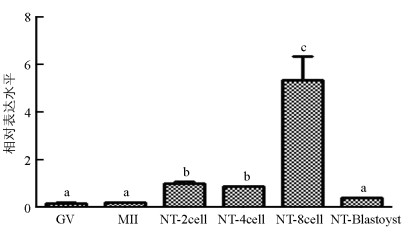
qRT-PCR结果显示, SMYD3基因在猪卵母细胞体外成熟过程中表达较低, 显著低于孤雌激活(囊胚除外)和核移植早期胚胎表达水平(p<0.05). SMYD3基因在MII期和GV(Germinal Vesicle)期表达水平差异不具有统计学意义(p>0.05);在孤雌激活早期胚胎中, SMYD3基因表达从2细胞期开始升高, 4细胞期最高, 随后降低, 呈现出先升高再降低的表达趋势(图 1). SMYD3基因在猪核移植早期胚胎中也呈现出先升高再降低的趋势, 4细胞期表达量低于2细胞期, 但差异不显著(p>0.05), 随后在8细胞期达到最高, 然后降低(图 2).
-
统计结果显示, 核移植胚胎的2细胞分裂率在各实验中差异不具有统计学意义(p>0.05). 上调SMYD3组核移植胚胎的4细胞率显著高于对照组[(45.6%±1.65%) vs (42%±0.56%), p<0.05], 下调组4细胞率与对照组差异不具有统计学意义(p>0.05). 与对照组相比, 上调SMYD3组核移植胚胎的囊胚率显著提高[(14.6%±0.96%) vs (9.6%±0.19%), p<0.05], 下调组囊胚率显著降低[(6.7%±0.71%) vs (9.6%±0.19%), p<0.05](表 3).
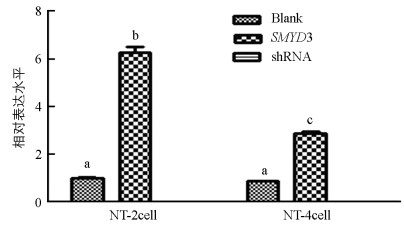
qRT-PCR结果显示, 与对照组相比, 上调组SMYD3基因表达水平在2细胞和4细胞期胚胎中显著提高(p<0.05), 下调组SMYD3基因检测不出. 上调组2细胞期胚胎的SMYD3基因表达显著高于4细胞期(p<0.05)(图 3).
-
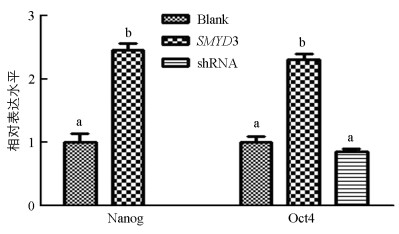
与对照组相比, 上调SMYD3组囊胚的Nanog和Oct4基因表达水平显著提高(p<0.05). 下调组囊胚中Nanog基因检测不出, Oct4基因表达低于对照组, 但差异不具有统计学意义(p>0.05)(图 4).
2.1. SMYD3基因在体外成熟猪卵母细胞和早期胚胎中的表达模式
2.2. 调控猪成纤维细胞SMYD3基因表达对体细胞核移植早期胚胎发育的影响
2.3. 调控SMYD3基因表达对猪核移植囊胚多能性基因表达的影响
-
近年来, 体细胞核移植产生的动物克隆效率低, 胚胎表型异常和生存能力低被认为是供体核的重编程不完全所致. 起初Bourc'his等[13]通过免疫荧光来跟踪染色体上的整体基因组甲基化变化, 结果显示在受精的牛体细胞核移植胚胎中植入前甲基化具有与自然繁殖的哺乳动物胚胎相似的模式, 但其甲基化动力学过程有着不同程度的紊乱. 而Dean等[14]对小鼠、猪和牛的比较研究中进一步印证了哺乳动物中甲基化的保守性与体细胞移植胚胎甲基化的异常. 之后研究者利用亚硫酸氢盐分析法对这种异常的甲基化进一步分析表明, 在牛核移植桑葚胚和囊胚与牛体外受精胚胎相比DNA甲基化水平较高, 水平与供体细胞DNA甲基化类似[15]. 同样在身为大动物的猪中, 研究者也发现与IVF(In Vitro Fertilization)早期胚胎相比, 发现组蛋白甲基化水平在核移植早期胚胎中异常[16], H3K36me3在猪体细胞核移植早期, 即胚胎1细胞时期不能被完全去甲基化[17]. 在核移植早期胚胎中DNA和组蛋白甲基化水平异常, 而体细胞重编程不完全造成了这些异常的甲基化水平.
多年研究表明, SMYD3能通过使染色体组蛋白H3K4发生二甲基化、三甲基化或抑制H4K5/H4K20的甲基化, 来改变染色质的可及性以及影响下游多种功能基因, 从而参与抑制肿瘤细胞凋亡、促进细胞增殖等生命活动过程. Bai等[10]发现过表达SMYD3基因可提高牛胎儿成纤维细胞iPSCs诱导效率, 证实其具有促进牛体细胞重编程的作用. 而下调SMYD3基因表达对斑马鱼、小鼠和牛胚胎发育具有不利影响, 证明SMYD3基因对胚胎发育有重要作用[9-12]. 因此, 本文先分析了SMYD3基因在猪卵母细胞体外成熟和早期胚胎发育过程中的表达模式, 发现在猪卵母细胞体外成熟过程中SMYD3基因表达无显著性差异, 这与前人对多数哺乳动物的研究相符合. 而体外成熟过程中SMYD3基因表达低于孤雌激活(囊胚除外)和核移植早期胚胎发育过程, 这种表达模式不同于牛来源细胞中的研究, 本文推测这种差异性来自于获得早期胚胎的方式不同, 或存在物种差异. SMYD3基因在孤雌激活和核移植早期胚胎发育中均呈现出先升高后降低的表达趋势, 却分别在4细胞期和8细胞期达到峰值, 说明SMYD3基因参与两者发育过程中的时间存在差异. 早期胚胎发育到4细胞至8细胞期, 正处于大批合子型基因转录时期, 这种状态将一直持续到早期囊胚;而8细胞期阶段胚胎正处于母型调控向合子型调控转变的时期, 也是在离体培养过程中需要克服发育阻断现象的时期, 涉及早期胚胎发育基因的适时激活, 瞬时表达、定位或消失等基因调控机制. Li等[18]对猪孤雌胚胎与体外受精胚胎发育过程中的染色质重编程进行比较, 发现孤雌胚胎在合子基因组激活的4细胞时期存在特殊的染色质区室解体过程. SMYD3在上述阶段的表达差异, 提示其在调控胚胎重编程的过程中应有重要作用.
Oct4基因在4~8细胞期被激活, 被认为是在早期发育过程中参与第1次细胞命运决定, 使细胞发育为滋养层或保留一部分全能细胞的关键因子. 第2次命运决定出现在囊胚期, 内细胞团部分保持亚全能性, 部分分化为原始胚层, Nanog在这时期起决定作用[19]. 两者作为多能性标志因子, 其表达水平对早期胚胎重编程具有重要作用. 前人研究表明, 下调SMYD3基因表达在小鼠IVF囊胚中抑制Nanog和Oct4等多能性基因表达[9]. Bai等[10]在牛IVF胚胎中下调SMYD3基因表达会出现发育阻滞的现象, 能显著促进8细胞期胚胎Nanog基因表达, 对OCT4基因无显著影响. 本实验检测了猪核移植囊胚Nanog和Oct4基因表达情况, 其中下调SMYD3组囊胚Nanog基因检测不出, 证明下调SMYD3基因可以抑制Nanog基因表达. Nanog基因表达水平较低难以检出, 与小鼠研究结果相符, 与牛研究结果不同, 推测原因可能是胚胎时期或来源不同所造成的结果差异;与对照组相比, 下调SMYD3组囊胚OCT4基因无显著差异, 与小鼠研究结果不同, 推测存在物种差异. 上调SMYD3基因表达促进囊胚中Nanog和Oct4基因表达, 初步说明SMYD3基因促进了猪核移植早期胚胎的重编程, 具有促进猪体细胞重编程的作用.
-
1) SMYD3基因在猪卵母细胞体外成熟过程中表达较低, 在孤雌激活和核移植早期胚胎发育过程中均呈现出先升高后降低的表达模式.
2) 上调SMYD3基因表达可提高猪体细胞克隆的囊胚发育率, 促进Nanog和Oct4基因表达提高核移植胚胎体外发育能力.





 DownLoad:
DownLoad: