-
开放科学(资源服务)标志码(OSID):

-
玉米(Zea mays L.)是我国广泛种植的粮饲作物,也是重要的生物能源和工业原料,在我国农业生产中占据重要地位[1]. 随着我国种植结构的调整和气候变暖导致的媒介昆虫大面积迁飞和分布,玉米矮花叶病、玉米粗缩病、玉米致死性坏死病等病毒病害在我国一些玉米种植区间歇性大面积发生. 目前在我国玉米上已检测出甘蔗花叶病毒(sugarcane mosaic virus,SCMV)、水稻黑条矮缩病毒(rice black streaked dwarf virus,RBSDV)、玉米褪绿斑驳病毒(maize chlorotic mottle virus,MCMV)、玉米黄花叶病毒(maize yellow mosaic virus,MaYMV)、留尼旺玉米线条病毒(maize streak Reunion virus,MSRV)等12种病毒,严重威胁着我国玉米的生产[2]. 甘蔗(Saccharum officinarium L.)在四川、云南、广东和福建等地区广泛种植,是我国最重要的糖料作物,也是生产葡萄糖、乙醇等产品的重要原料. 甘蔗上的常发病毒病主要包括SCMV、高粱花叶病毒(sorghum mosaic virus,SrMV)和甘蔗线条花叶病毒(sugarcane streak mosaic virus,SCSMV)引起的甘蔗花叶病,甘蔗黄叶病毒(sugarcane yellow leaf virus,SCYLV)引起的甘蔗黄叶病,以及甘蔗线条病毒(sugarcane streak virus,SSV)、甘蔗斐济病病毒(sugarcane Fiji disease virus,SFDV)和甘蔗杆状病毒(sugarcane bacilliform virus,SCBV)等引起的病毒病[3].
SCMV在我国广泛分布,其引起的玉米矮花叶病和甘蔗花叶病是我国玉米和甘蔗上的两种重要病毒病害[4-5]. SCMV是马铃薯Y病毒科(Potyviridae)马铃薯Y病毒属(Potyvirus)的成员,经由玉米蚜、麦二叉蚜等多种蚜虫以非持久性方式进行传播,是侵染玉米、高粱、甘蔗等禾本科植物的重要病毒病原[6-8]. SCMV侵染可诱导花叶、褪绿和矮化等多种症状,最终造成严重的产量损失[9-10]. SCMV为线状病毒粒体,包裹1条长约10 000碱基且3′末端有poly(A)结构的正单链RNA基因组[11]. SCMV基因组编码1个大的多聚蛋白,随后被切割为10个成熟蛋白以及转录滑移所产生的P3N-PIPO蛋白[12]. 马铃薯Y病毒属病毒编码的外壳蛋白(CP)是一个多功能蛋白,参与病毒基因组的包被、病毒移动、蚜虫传播等生物学过程[13-14]. 因为外壳蛋白基因功能和序列的保守性,通常被用于病毒分类和病毒变异、重组等遗传进化研究[8, 15].
病毒检测和病毒群体的遗传变异分析有利于了解病毒的地理分布和寄主适应性,且有益于对病毒病流行趋势进行评估,是病毒病害防控的重要手段[16-18]. 玉米是我国西南地区广泛种植的重要粮饲作物,甘蔗是四川和云南部分地区种植的重要经济作物,但病毒病的发生严重威胁着玉米和甘蔗的生产. 基于本课题组前期对四川玉米和甘蔗病毒病田间调查结果,本研究从四川攀枝花市采集共计54份表现花叶、斑驳、矮化等症状的玉米病叶和42份甘蔗病叶,以小RNA高通量测序结合反转录-聚合酶链式反应(reverse transcription-polymerase chain reaction,RT-PCR)法对玉米和甘蔗病样中的病毒种类和发生率进行检测;并扩增获得四川玉米上6个和甘蔗上5个SCMV分离物的CP基因序列,综合NCBI数据库中的SCMV所有中国分离物CP基因序列分析SCMV四川分离物群体的遗传变异情况.
HTML
-
分别于2019年7月和2020年8月从四川攀枝花田间采集表现花叶、褪绿、斑驳、矮化等症状的玉米病叶样品54份和甘蔗病叶42份,保存于-80 ℃冰箱中.
-
克隆载体pMDTM19-T,日本TaKaRa公司;大肠杆菌Escherichia coli Trans5α,北京全式金生物技术有限公司.
-
Ex Taq? DNA聚合酶,RNAiso Plus总RNA提取试剂,M-MLV反转录试剂盒,T4 DNA连接酶,日本TaKaRa公司;通用型DNA纯化回收试剂盒,天根生化科技(北京)有限公司.
-
本研究中所用引物序列信息见表 1,SCMV检测引物引自文献[19],其他检测引物和SCMV CP基因扩增引物根据GenBank中多个病毒分离物基因组序列多重比对分析后在序列保守区设计.
-
取玉米或甘蔗病叶约0.1 g,充分研磨后用TaKaRa公司RNAiso Plus试剂参照说明书流程提取总RNA,-80 ℃冰箱中保存备用.
以提取的总RNA为模板,用TaKaRa公司的M-MLV反转录试剂盒进行反转录合成cDNA. 反应体系(10 μL):总RNA 1 μg,Oligo(dT) Primer (50 μmol/L) 1 μL,Random Primers (25 μmol/L) 1 μL,RNase-free ddH2O补至6 μL;于70 ℃水浴孵育10 min后冰上冷却2 min;向上述溶液中依次加入5×M-MLV buffer 2 μL,10 mmol/L dNTP混合物0.5 μL,40 U/μL RNA酶抑制剂0.25 μL,200 U/μL M-MLV反转录酶0.5 μL,RNase-free ddH2O补至10 μL;42 ℃保温1 h. 反应获得cDNA产物直接用于PCR扩增或于-20 ℃冰箱中保存备用.
以反转录获得cDNA为模板进行PCR扩增,病毒检测PCR反应体系(25 μL):ddH2O 11.0 μL,cDNA 0.5 μL,10 μmol/L上下游引物各0.5 μL,PCR mix 12.5 μL. SCMV CP基因克隆的PCR反应体系(50 μL):ddH2O 37.5 μL,10×Ex Taq buffer(Mg2+ plus)5.0 μL,dNTPs 4.0 μL,cDNA 1.0 μL,10 μmol/L上下游引物各1.0 μL,Ex Taq 0.5 μL. PCR反应程序设定:94 ℃预变性5 min;94 ℃变性30 s后退火30 s(退火温度根据引物熔解温度确定),72 ℃延伸30~60 s(根据扩增片段长度确定),34次循环;最后72 ℃延伸10 min. PCR产物经1%琼脂糖凝胶电泳分析.
-
PCR产物用天根通用型DNA纯化回收试剂盒(Universal DNA Purification Kit,TIANGEN)参照说明书流程纯化回收. 回收产物连入pMDTM19-T载体,连接反应体系(10 μL):10×T4 DNA Ligase buffer 1 μL,回收产物5 μL,pMDTM19-T 0.5 μL,T4 DNA Ligase 0.5 μL,ddH2O补至10 μL,置于16 ℃过夜反应. 将连接产物转化大肠杆菌Trans5α感受态,37 ℃培养后挑取单菌落在含抗生素(100 mg/L氨苄青霉素)LB液体培养基中培养6~8 h,菌液经PCR检测后选阳性克隆进行测序分析.
-
经PCR检测呈阳性的菌液委托生工生物工程(上海)股份有限公司测序,测序结果在NCBI(National Center for Biotechnology Information)数据库用BLAST进行序列相似性检索. 用本研究中克隆获得SCMV四川分离物CP基因和NCBI数据库中已上传的所有SCMV中国分离物CP基因序列进行系统进化分析,以玉米矮花叶病毒(maize dwarf mosaic virus,MDMV)CP基因为外组,用MEGA4软件的邻接(Neighbor-Joining)法构建系统进化树,重复取样1 000次[20]. SCMV核苷酸变异、群体分化和基因流、群体中性测试均用DNaSP软件进行分析[21].
1.1. 材料
1.1.1. 毒源
1.1.2. 载体和菌株
1.1.3. 主要试剂
1.1.4. 引物
1.2. 方法
1.2.1. 总RNA提取及RT-PCR
1.2.2. DNA纯化回收、连接T载体及转化大肠杆菌
1.2.3. 序列测定及生物信息学分析
-
从四川攀枝花田间采集表现病毒病症状的玉米和甘蔗植株病叶,玉米叶片主要表现褪绿、斑驳和花叶症状,甘蔗叶片主要表现褪绿、黄化、坏死和叶卷曲症状(图 1).
为综合分析所采集样品中存在的病毒种类,各样品均取0.1 g混合研磨制备混样并提取总RNA,委托上海美吉生物医药科技有限公司进行小RNA高通量测序. 测序共获得13 868 023个reads,去除接头以及长度小于18或大于32个核苷酸的reads,获得9 505 185个Clean reads. 将Clean reads通过从头组装产生重叠群后与病毒数据库GenBank Virus RefSeq进行比对并注释得到的病毒序列. 所有重叠群共比对到5种病毒:SrMV,MaYMV,SCMV,SCBGAV和MSRV(表 2).
为验证玉米和甘蔗样品中存在的病毒种类,分别提取本研究中采集的54份玉米和42份甘蔗样品总RNA,反转录合成cDNA,以表 1中引物分别进行PCR扩增,琼脂糖凝胶电泳检测结果表明:四川攀枝花玉米病样中可检出SCMV,MaYMV和MSRV 3种病毒,检出率分别为66.7%,59.3%和3.7%;甘蔗病样中可检出SCBGAV,SCMV和SrMV 3种病毒,检出率分别为76.2%,52.4%和28.6%(表 3). 从每种病毒的阳性样品中随机选取2个PCR扩增产物经切胶回收后连入pMDTM19-T载体,转化大肠杆菌Trans5α感受态,挑取阳性克隆进行序列测定,序列分析显示扩增产物均为对应病毒基因组序列,表明扩增出目标片段的玉米或甘蔗样品被该病毒侵染.
-
目前GenBank数据库中尚无SCMV四川分离物的基因序列,本研究随机选取6个检出SCMV的玉米和5个检出SCMV的甘蔗样品cDNA为模板,以引物对SCMV-CP-F/SCMV-CP-R扩增CP基因全序列. 扩增产物经纯化回收后连入pMDTM19-T载体并转化大肠杆菌Trans5α感受态,每个转化子挑取3个阳性克隆经测序鉴定SCMV CP基因序列(表 4).
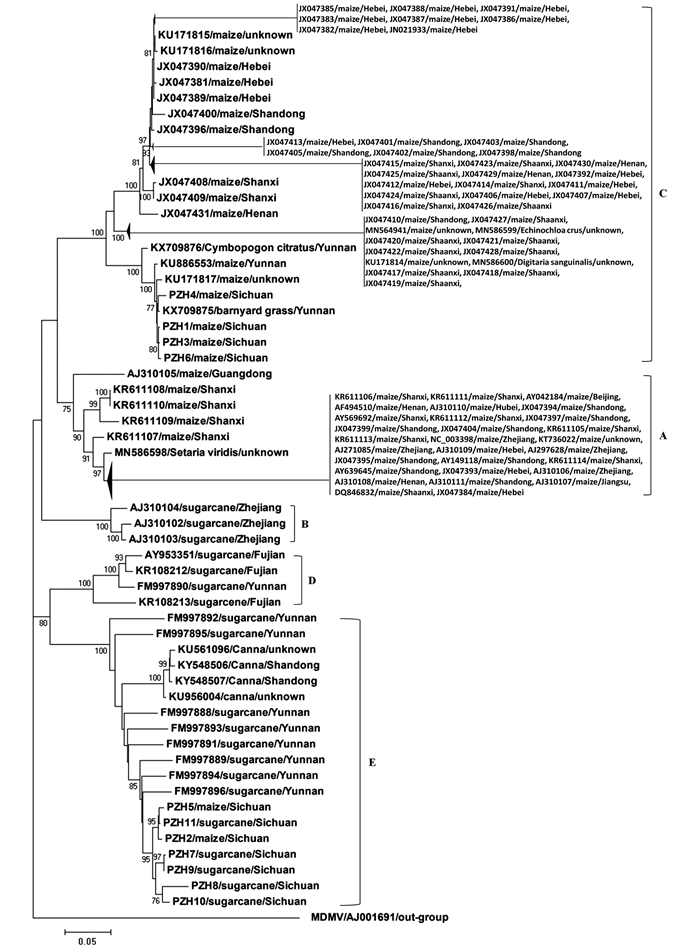
为综合分析SCMV四川分离物群体在SCMV中国分离物群体中的遗传进化,利用从四川玉米和甘蔗上扩增获得的SCMV分离物CP基因序列,并从GenBank数据库中下载获得所有来源于我国不同地区及寄主的SCMV CP基因序列,构建系统进化树并分析其遗传变异情况. 序列比对结果显示,120个SCMV中国分离物的CP基因序列的核苷酸同源性为73.8%~100.0%. 以MEGA4软件构建系统进化树,结果显示,所有SCMV中国分离物聚为5个进化组分支(图 2).
A组和C组中SCMV分离物主要来自玉米,而B组和D组分离物则主要来自甘蔗. 来自于玉米的SCMV四川分离物PZH1,PZH3,PZH4和PZH6位于C组,而来自于甘蔗的SCMV四川分离物PZH7,PZH8,PZH9,PZH10和PZH11均位于E组,表明SCMV的遗传进化具有明显的寄主特异性. 进化树聚类结果还表明,来自于四川和云南甘蔗上的SCMV分离物主要聚于E组,来自于福建和浙江甘蔗上的SCMV分离物分别聚于D组和B组,来自于四川、山西、陕西、河北和山东几个地域依次毗邻省份玉米上的SCMV分离物同聚于C组,即来自于同种寄主、同一省份或邻近省份的SCMV分离物可聚为同一组,这说明地理起源对于SCMV的遗传进化也具有重要作用.
以DnaSP5软件计算SCMV中国分离物群体的核苷酸多样性为0.129 88,表明SCMV中国分离物群体的总体变异度较低. 对SCMV分离物序列信息超过10个的四川、云南、河北、山西、陕西、山东等地的SCMV分离物CP基因的核苷酸多样性进行分析,结果表明,来源于河北、山西和陕西3省的SCMV分离物核苷酸变异度更低,且均来自于各省相同的宿主. 而来源于四川、云南和山东的SCMV分离物来自于至少两种以上的宿主,且各省SCMV群体的核酸变异度也相对更高(表 5). 该结果与对SCMV系统进化的分析一致,均可表明宿主在SCMV中国分离物的遗传分化过程中发挥着重要的作用.
基于上述结果分析,宿主和地理分布在SCMV中国分离物的遗传进化过程中作用明显,且SCMV在田间主要由蚜虫进行传播,暗示着邻近地理区域的SCMV群体可能因蚜虫的交叉传播出现趋同进化. 本研究以DnaSP5软件分析了四川与云南、山西、陕西、河北和山东等省SCMV分离物群体间的遗传分化和基因流,种群间的遗传分化程度采用Ks*,Z和Snn 3个参数来反映,种群间的基因流采用FST值来反映[16, 21-22]. FST测试结果表明,SCMV四川和云南分离物之间的FST值(表示不同群体间的遗传多样性,反映基因流迁移率)为0.011 23,而四川与其他省份分离物群体间的FST值均大于0.33,表明四川和云南的SCMV分离物群体间基因流更为频繁(表 6). 本研究中的SCMV四川分离物来源于四川最南端川滇结合部的攀枝花市,与云南相邻,这表明更近的地理分布更易导致相邻地区SCMV群体的频繁基因流.
SCMV四川和云南分离物间的频繁基因流暗示这两个区域间的SCMV可能以某种方式(如蚜虫的田间传播)进行着交叉传播. 根据SCMV中国分离物CP基因系统进化树分组结果,SCMV四川和云南分离物均主要聚于C组和E组. 为分析C组和E组的SCMV四川和云南分离物种群(分别表示为SC-YN-GpC和SC-YN-GpE)的扩展态势,本研究通过Tajima's D,Fu and Li's D和Fu and Li's F等方法进行中性测试[23-24]. 结果显示,来自于C组和E组的四川和云南分离物群体的Tajima's D,Fu and Li's D和Fu and Li's F值均为负值,但未达到统计学意义,表明SC-YN-GpC和SC-YN-GpE两个种群在四川和云南可能存在扩张趋势(表 7).
2.1. 四川玉米和甘蔗病样中的病毒检测
2.2. SCMV四川分离物CP基因克隆及遗传变异分析
-
本研究是对四川攀枝花玉米和甘蔗中病毒病原种类和侵染率的综合分析,也是对SCMV四川分离物群体遗传变异情况的首次报道. 我国玉米和甘蔗上的病毒病害长期发生,随着病毒鉴定技术的发展,目前我国已在田间玉米和甘蔗上鉴定到多种病毒[2-3, 25-26]. 本研究从四川攀枝花市玉米上检出SCMV,MaYMV和MSRV 3种病毒,从甘蔗上检出SCBGAV,SCMV和SrMV 3种病毒,其中SCMV是引起该地区玉米和甘蔗病毒病的重要病原.
前期研究对世界上某些地区SCMV分离物的遗传进化分析结果暗示宿主和地理因素对SCMV进化的重要性[16, 27]. 本研究对中国不同省区SCMV分离物群体的遗传进化分析结果显示,SCMV中国分离物可聚为5个进化分支,且其系统进化和核苷酸多样性分析结果均表明,宿主和地理起源在SCMV的遗传进化过程中发挥着重要的作用. FST测试表明来源于川滇交界处的四川攀枝花SCMV分离物和云南分离物间存在频繁基因流,这也表明地理因素在SCMV遗传进化过程中同样作用明显,这可能与蚜虫在较近的地理距离内频繁交叉传播SCMV有关. 本研究中两个来源于玉米的分离物PZH2,PZH5和来源于四川甘蔗的5个分离物聚入同一组,这可能也是由于蚜虫在玉米和甘蔗间交叉传播SCMV所导致.






 DownLoad:
DownLoad: