-
开放科学(资源服务)标志码(OSID):

-
结球甘蓝在我国广泛种植,是十字花科芸薹属重要的蔬菜作物之一[1]. 黑腐病是为害甘蓝生产的主要细菌性病害之一,病原菌为野油菜黄单胞菌野油菜致病变种(Xanthommonas campestris pv. campestris),简称为“Xcc”[2]. 黑腐病病原菌为水孔侵入,侵染子叶时呈水浸状,然后迅速蔓延至真叶[3];侵染真叶时,病菌多从叶缘侵入,病斑从叶缘的褪绿斑点逐渐向叶片的主脉扩张,形成“V”形病斑,颜色从黄色逐渐变为褐色[3-4]. 黑腐病病菌也可从伤口侵入,在叶侵染部位形成不规则的淡褐色病斑[5]. 近年来,黑腐病在我国甘蓝主产区普遍发生,直接为害甘蓝的叶片和叶球,对甘蓝生产造成严重为害[6]. 尽管可通过轮作换茬、喷施药物防护等方式加强田间管理以预防黑腐病的发生,但通过农业措施防治黑腐病效果微弱且会增加管理成本,而药物防治又具有容易产生抗药性、增加环境污染和农药残留等问题[7]. 因此,种植抗黑腐病甘蓝品种成为防治该病最经济有效的措施.
黑腐病病原菌有11个生理小种[8-9],其中,Xcc1和Xcc4是世界上黑腐病发生的主要生理小种[4, 10-11]. 甘蓝中存在少量抗Xcc1的种质[12-13],但是抗Xcc1和Xcc4的资源相对较少[14-15]. 然而,甘蓝的野生种和近缘种中存在大量抗黑腐病的资源. 研究发现,甘蓝野生种Brassica montana “UNICT5169”和Brassica balearica “PI435896”高抗Xcc4,与花椰菜杂种F2的抗性表现为数量性状遗传[16]. 埃塞俄比亚芥“NPC-9”抗Xcc1,“PI199947”抗Xcc1和Xcc4,At1g70610标记可以用于其与花椰菜杂种的黑腐病抗性辅助鉴定[10, 15, 17-18]. Bhatia等[19]发现埃塞俄比亚芥Xcc1和Xcc4生理小种的抗性存在多基因遗传,在Brassica oleracea × Brassica carinata的小孢子培养后代中,所有连锁群为B7和B5的植株均具有对Xcc1的抗性,连锁群为B6和B2的杂种后代则具有对Xcc4的抗性,而缺乏这两个连锁群的株系对Xcc4表现为易感. Tonguç等[20]在芥菜中也发现抗Xcc1和Xcc4的种质“A19182”和“A19183”,其与甘蓝的杂种F1和BC1抗Xcc1和Xcc4. 在抗黑腐病甘蓝(“SCNU-C-3470”) C08染色体上发现了抗病基因Bol031422,BR6-InDel标记可用于Xcc6和Xcc7的抗性辅助选择[14]. 这些丰富的资源和分子标记为甘蓝黑腐病的抗性鉴定和改良提供了重要的支撑.
翟文慧[21]曾通过鉴别寄主法对我国11个地区的十字花科蔬菜黑腐病菌的生理小种分化进行了研究,发现我国黑腐病病原菌大部分为Xcc1和Xcc4,西南地区的十字花科黑腐病病原菌主要是Xcc1,但尚未发现利用分子标记辅助鉴定重庆地区黑腐病病原菌生理小种的相关研究. 另外,抗性鉴定是筛选抗黑腐病材料十分重要的环节,甘蓝黑腐病抗性鉴定体系主要采用苗期接种法和离体整叶接种法. 针对种子量极少或植株长势差的材料,苗期接种法和离体整叶接种法的操作性受限,可是采用离体叶片滴接法和喷雾法能快速实现甘蓝黑腐病的苗期鉴定[22]. 本研究首先采用分子标记辅助鉴定重庆地区黑腐病病原菌生理小种,再将其用离体叶片喷雾法接种于30份结球甘蓝材料上[23],筛选出抗黑腐病的甘蓝材料,然后利用叶片打孔滴接法和打孔喷雾法,对甘蓝黑腐病抗性进行验证,以期建立一套高效快捷的甘蓝黑腐病抗性鉴定方法,为甘蓝抗黑腐病的种质创新和新品种选育提供技术保障.
HTML
-
甘蓝黑腐病菌(Xanthommonas campestris pv. campestris),为黄单胞杆菌属,甘蓝黄色杆菌,是重庆菌株,由西南大学植物保护学院提供.
-
由西南大学十字花科蔬菜研究所提供的30份结球甘蓝材料,以感病品种“西园四号”作为对照.
-
用平板划线法将甘蓝黑腐病菌株接种于NA培养基上,在28 ℃恒温培养箱中培养48 h后取出,挑选其中的单菌落用平板划线法再次接种于新的NA培养基上,置于28 ℃恒温培养箱中培养48 h,重复3次. 挑选最后一次得到的平板上的单菌落转管并封装,放置于-80 ℃冰箱中保存备用. 进行接种试验之前,将封装保存于-80 ℃冰箱中的甘蓝黑腐病菌株取出,用NA培养液培养并在摇床上摇菌24 h,后利用血球计数板将菌株制备成浓度为1.0×108 cfu/mL的菌悬液,放置在4 ℃冰箱中保存备用[23].
-
于甘蓝莲座期前,选取30份结球甘蓝和对照植株从里到外数的第3片叶,贴上标签,其中每份材料选取30个单株,用于整叶喷雾法鉴定[24]. 在培养架上放置湿毛巾,将离体叶片置于湿毛巾上,将提前准备好的菌液装入喷壶中,均匀喷施于叶片表面,然后用塑料薄膜覆盖,四周用夹子和砖块固定,使其内部保持高湿状态,防止水分蒸发. 调节接种室内温度至25 ℃,给予材料最佳的发病环境. 每隔2 d观察一次并拍照,至叶片完全枯萎时统计病斑大小,筛选抗性材料.
-
根据整叶喷雾法筛选出极端抗黑腐病的结球甘蓝材料,随机选取3个单株的3片叶片,使用直径1.4 cm孔径的打孔器在每片叶子上对称打孔20个,形成叶盘. 将90 mm培养皿洗净并垫上滤纸保湿,每皿放置10个叶盘. 用装有菌液的喷壶均匀喷施叶盘,盖好培养皿,放置于保鲜盒中,置于25 ℃接种箱中,5 d后统计病斑大小.
-
将每片叶片剩下的10个叶盘放置于培养皿中,使用移液枪滴20 μL菌液于叶片中央. 盖好培养皿,放置于保鲜盒中,置于25 ℃接种箱中,5 d后统计病斑大小.
-
1) 测量整叶或叶盘长度,病斑的发病点到病斑末端的长度,病斑大小=病斑长度/叶片长度×100%. 同时按黑腐病抗性分级标准[23-25]进行分级.
Ⅰ病情分级标准.
0级:没有病斑;
1级:叶片有黑色枯死点,无扩展;
3级:枯死点向外扩展,25%以下;
5级:枯死点向外扩展,25%~50%;
7级:枯死点向外扩展,50%~75%;
9级:枯死点向外扩展,75%以上.
病情指数=100×∑(各级病叶数×各级代表值)/(调查总叶数×最高级代表值).
Ⅱ抗性分级标准.
免疫(Ⅰ):病情指数0.00;
高抗(HR):0<病情指数≤11.11;
抗病(R):11.11<病情指数≤33.33;
耐病(T):33.33<病情指数≤55.55;
感病(S):55.55<病情指数≤67.77;
高感(HS):67.77<病情指数≤100.
考虑到离体整叶喷雾法发病条件极端,需要对其抗性分级标准进行调整,以便适应株系的真实抗性,调整后如下.
免疫(Ⅰ):病情指数0.00;
高抗(HR):0<病情指数≤22.22;
抗病(R):22.22<病情指数≤44.44;
耐病(T):44.44<病情指数≤77.77;
感病(S):77.77<病情指数≤100.
2) 分子标记辅助选择
重庆本土黑腐菌的分子标记鉴定采用Rubel等[26]以及Afrin等[27]的方法,略作修改. 其中,Xcc-47R1(1 089 bp)鉴定Xcc1生理小种,XccR3-49(867 bp)和XccR3-52(1 889 bp)鉴定Xcc3生理小种,Xcc1-46R4(462 bp)和Xcc2-46R4(578 bp)鉴定Xcc4生理小种. 甘蓝黑腐病抗性采用来自埃塞俄比亚芥的黑腐病抗性标记(Black rot 3,Black rot 6)(本实验室开发)[28]和甘蓝中的分子标记(BR6-InDel[14])进行分子标记辅助选择. 具体引物信息见表 1.
-
本研究所用的t-test 和相关性分析均采用SPSS 软件进行处理.
1.1. 试验材料
1.1.1. 菌种来源
1.1.2. 甘蓝材料来源
1.2. 试验方法
1.2.1. 菌种纯化与菌悬液制备
1.2.2. 整叶喷雾法
1.2.3. 打孔喷雾法
1.2.4. 打孔滴接法
1.2.5. 抗性鉴定
1.2.6. 数据分析
-
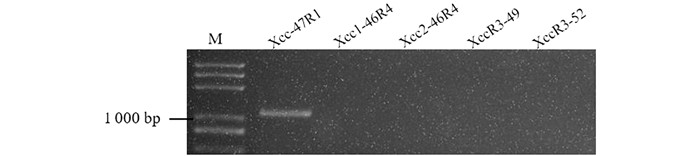
前期研究发现黑腐病的主要流行病原菌是Xcc1和Xcc4生理小种[10],本研究采用的黑腐菌是重庆本地菌种,为了明确使用的黑腐病病原菌生理小种,本研究利用能够区分黑腐菌Xcc1 (Xcc-47R1,1 089 bp),Xcc3 (XccR3-49,867 bp;XccR3-52,1 889 bp)和Xcc4 (Xcc1-46R4,462 bp;Xcc2-46R4,578 bp)生理小种的分子标记对黑腐菌进行分子标记辅助鉴定. 结果发现,除了Xcc-47R1能够扩增出约1 089 bp的特异条带,其他引物扩增均无条带,表明使用的重庆本地黑腐病病原菌是Xcc1生理小种(图 1).
-
离体叶片喷雾法是甘蓝黑腐病抗性鉴定广泛使用的方法之一,本研究首先采用离体叶片整叶喷雾法对30份结球甘蓝材料和对照进行黑腐病抗性鉴定. 为了更有效地筛选出抗黑腐病的结球甘蓝材料,本研究采用极端抗性筛选法,对接种后16 d的叶片进行黑腐病发病情况的统计. 结果发现,2份材料的平均病斑大小低于30%,3份材料的平均病斑大小介于30%和60%之间,其余25份材料的平均病斑大小都高于60%. 其中,S18(24.80±19.70%),S18S(28.57±0.01%)和S232(43.18±9.64%)的平均病斑大小显著小于对照(87.04±18.33%),其余材料与对照没有显著性差异. S18的病情指数为38.89,表现为抗病. S18S(55.55),S232(66.67),S23S(71.72),167(74.91),165(76.30)和169(76.30)表现为耐病,其余23份材料与对照一样为感病材料(图 2、表 2).
-
尽管采用离体叶片喷雾法,极端处理16 d可筛选出抗黑腐病的材料,但是该过程周期长,所需叶片多,针对植株少的材料无法准确地鉴定. 因此,本研究试图比较打孔喷雾法和打孔滴接法,快速有效地鉴定出抗黑腐病的材料. 本研究根据黑腐病极端抗性鉴定结果,挑选1份抗病材料(S18)、1份耐病材料(S232)和2份感病材料(S19、S22)与对照(西园四号)一起比较离体叶片打孔喷雾法和打孔滴接法的鉴定效果. 每个材料选取3片大小一致的叶片,每个叶片打孔产生20个叶盘,其中10个叶盘进行喷雾接菌,10个叶盘进行滴接接菌,5 d后统计发病情况. 滴接法显示S18(DI=31.11)为抗病,S232(DI=44.44)为耐病,S19,S22和对照均表现高感,并且打孔滴接法与离体叶片整叶喷雾法病情指数显著正相关(r=0.96,p=0.011). 喷雾法显示S18(DI=39.68)和S232(DI=53.97)为耐病,S19,S22和对照均表现高感(图 3、表 3). 尽管打孔喷雾法无法区分出抗病和耐病的材料,但是打孔喷雾法与离体叶片整叶喷雾法显著正相关(r=0.97,p=0.006),与打孔滴接法的病情指数也显著正相关(r=0.99,p=0.002). 该结果说明3种方法都能进行黑腐病的抗性鉴定,打孔滴接法能更准确地区分抗病材料和耐病材料.
-
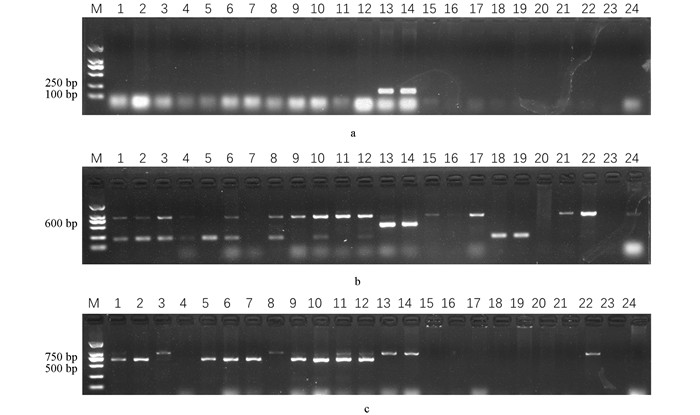
由于前期研究发现大量利用埃塞俄比亚芥与甘蓝杂交的相关研究,本研究筛选出的甘蓝种质是否含有埃塞俄比亚芥的黑腐病抗性成分尚不明确. 为了探究筛选出甘蓝种质黑腐病抗性标记,本研究利用前期从抗黑腐病埃塞俄比亚芥(PI199947和PI199949)中鉴定的抗黑腐病连锁标记(Black rot 3,250 bp和Black rot 6,500 bp)对抗病材料S18的6个后代和耐病材料S232的6个后代以及10个感病甘蓝材料进行分子标记辅助选择. 结果发现,除了埃塞俄比亚芥抗性材料(PI199947和PI199949)外,其余材料均无目标条带. 但是,利用与Xcc6 / Xcc7连锁的分子标记BR6-InDel(抗病条带724 bp,感病条带1 013 bp)进行鉴定,发现尽管所有感病材料中均不含有抗性标记,但是S18的后代中有3个单株只含有抗性条带,1个单株含有抗/感条带;S232的后代中有1个单株只含有抗性条带,4个单株含有抗/感条带(图 4),说明该标记可能对于耐病材料能够进行分子辅助选择,但是无法区分抗病和耐病材料. 该研究结果表明S18的黑腐病抗性可能不同于埃塞俄比亚芥,需要开发新的分子标记.
2.1. 黑腐病病原菌分子标记鉴定
2.2. 利用离体叶片整叶喷雾法鉴定抗黑腐病材料
2.3. 利用离体叶片打孔法鉴定抗黑腐病材料
2.4. 黑腐病分子标记辅助鉴定
-
本研究利用黑腐菌喷施离体整叶极端处理16 d后筛选出抗黑腐病材料1份(S18),耐病材料6份(S18S,S232,S23S,165,167,169). 选取1份抗病(S18)、1份耐病(S232)和2份感病材料用于比较离体叶片打孔滴接法和喷雾法,结果发现,打孔滴接法可快速进行结球甘蓝的黑腐病抗性鉴定,筛选出抗黑腐病的材料.
-
本研究采用分子标记进行黑腐菌PCR鉴定,发现重庆本土菌种为Xcc1生理小种. 抗病甘蓝材料不含有埃塞俄比亚芥黑腐病抗性连锁标记(Black rot 3和Black rot 6),但是在抗病和耐病甘蓝后代中均存在BR6-InDel标记.
-
要建立高效的甘蓝抗病性筛选鉴定方法,既要符合田间发病期病原菌的适宜发病环境,也要考虑甘蓝植株自身生长状况与抗病性关系;并要进行接种鉴定选用致病力较强的病原菌,在甘蓝幼苗的旺盛生长期进行接种试验,才能真实地表现出甘蓝资源的抗感性能. 对黑腐病抗性鉴定常用接种方法有幼苗期喷雾接种、幼苗期剪叶接种、针刺接种、叶脉接种、水孔接种、吐水法和离体叶片法等10余种接种方法[29]. 不同的接种方法,由于叶片侵染时的状态、环境温湿度的不同,造成发病时间呈现较大的差异. 种子侵染接种较受国外学者青睐,但此法条件难以控制,受影响因素也较多. 陈果等[30]采用幼苗期喷雾接种,发现绝大多数材料的抗病性鉴定结果与田间抗性鉴定结果一致,但种质种子量较少的情况下该方法不适用. 幼苗期剪叶接种和喷雾接种使病原菌在水孔处繁殖,通过水孔侵入,不仅能鉴定寄主抗扩展能力,还能反映抗侵入特性,较剪叶法更能全面地反映寄主所具有的抗病性,但常出现菌体不能进入水孔的现象,造成感病品种也不发病. 吐水法接种可以避免漏接现象,而且鉴定结果较为准确,但费时费力,接种难度比较大[24]. 离体叶片接种易操作,不受季节时间限制和外界环境影响,但发病时间较长. 王超等[31]在甘蓝黑腐病研究中对比了5种接种方法,认为离体剪叶法更适用于甘蓝黑腐病的苗期鉴定. 本研究首先采用离体整叶喷施法进行极端处理,这种方法可同时筛选多份材料,在不损伤叶片的情况下得到的结果也更符合甘蓝的田间栽培环境,筛选出的一定为抗黑腐病材料,但是该方法耗时长,工程量大,筛选出来的结果比较极端,可能会筛掉一部分高抗和耐病品种. 因此,在总结以上接种方法后,本研究筛选出了一套简单迅速且鉴定准确的甘蓝黑腐病抗性鉴定方法——打孔接种法,分别为打孔喷雾接种法和打孔滴接法. 两种接种方法统计时间一致,共性在于同一方法不同材料之间的发病状况差异均较大,可以明显比较不同材料抗病性大小;而两种方法之间平行比较存在发病状态的差异. 在叶片破坏程度与叶片自身抗性等同的情况下,打孔喷雾法相较于打孔滴施法发病更快,但是这两种方法与整叶喷雾法的发病趋势均一致. 接种5 d后统计发病情况,打孔滴接法能区分抗感材料;打孔喷雾法可能适合提前统计. 因此,采用离体叶片打孔滴接法接种5 d后进行黑腐病发病情况的统计,可快速并可靠地进行结球甘蓝的黑腐病抗性鉴定.
挖掘和筛选抗源材料是开展甘蓝黑腐病育种工作的基础,国内外许多育种工作者已经在抗源筛选及新品种的选育上取得了一定的进展[32]. 但由于黑腐病生理小种分化较为复杂,单个抗性品种/品系并不能为Xcc所有的生理小种提供抗性. 因此,在分子水平上正确鉴定Xcc小种抗性位点对于黑腐病抗性育种以及利用现有抗性种质是必要的. 本研究利用Xcc1 (Xcc-47R1,1 089 bp),Xcc3 (XccR3-49,866 bp;XccR3-52,1 888 bp)和Xcc4 (Xcc1-46R4,462 bp;Xcc2-46R4,578 bp)的分子标记对重庆本土病菌鉴定后,发现重庆本土黑腐菌为Xcc1. 研究认为,十字花科A基因组抗Xcc4,B基因组抗Xcc1和Xcc4 [15],含有BBCC基因组的埃塞俄比亚芥可以同时高抗Xcc1和Xcc4 [17]. 近年来,在甘蓝中也发现了抗Xcc1的种质[12-13]. 本研究从甘蓝资源中筛选出抗Xcc1的材料,并利用前期从抗黑腐病埃塞俄比亚芥中筛选出的连锁标记Black rot 3和Black rot 6,对筛选出的抗黑腐病甘蓝材料后代进行分子标记辅助选择,发现在所有材料中均不含有埃塞俄比亚芥抗Xcc1的抗性位点,说明这些结球甘蓝存在不同于埃塞俄比亚芥的新Xcc1抗性位点;该研究对甘蓝黑腐病抗性基因的挖掘和抗性改良提供了重要的种质资源. Hong等[14]通过分析油菜芽孢杆菌中157个编码NBS-LRR的R基因,筛选到一个可能与Xcc6的黑腐病抗性紧密关联的基因Bol031422,从而开发了相关的InDel标记BR6-InDel,并且发现将接种Xcc6或Xcc7获得的表型结果与BR6-InDel标记的基因型结果进行比较,显示BR6-InDel标记适应性的预测能力为83.9%. 本研究将该标记应用到筛选出的抗病、耐病材料后代和感病材料中,结果发现在抗病和耐病材料中均能扩增出抗性条带,但是感病材料中无抗性条带,说明该标记并不能对S18进行黑腐病的抗性辅助选择,但是抗病材料(S18)和耐病材料(S23)可能具有Xcc6或Xcc7抗性.







 DownLoad:
DownLoad: