-
猪繁殖与呼吸综合征病毒(porcine reproductive and respiratory syndrome virus,PRRSV)主要导致母猪出现繁殖障碍和生长保育猪出现呼吸系统障碍疾病[1]. PRRSVs 20世纪80年代在美国首次出现,1991年Wensvoort等[2]在欧洲首次分离到该病毒LV(Lentivirus)疾毒株,此后PRRSV全球流行. 1996年我国兽医学家郭宝清等[3]在流产的猪胎中首次分离到PRRSV病毒CH-1a,证明了中国大陆存在猪繁殖与呼吸综合征病毒感染. 2007年,Tian等[4]报道了导致2006年我国爆发高致病性蓝耳病,以引起成年猪高热,耳朵发蓝,严重呼吸障碍,高死亡率为特征的疾病,对我国养猪业造成严重损失的HP-PRRSV类型毒株JXA1,此后我国分离到多株该类型毒株如HuN4[5-6].近年来,由于猪繁殖与呼吸综合征病毒具有广泛持续的遗传变异多样性特征,特别是高致病性毒株新亚型和流行新毒株的不断出现,给规模化猪场猪繁殖与呼吸综合征的防治带来了严重挑战,导致该病广泛流行给养猪业造成严重危害[7-9].目前主要靠疫苗免疫如HP-PRRSV毒株的JXA1,HUN4,TJ和经典的CH-1a疫苗对该病进行防治.但是大量研究表明,现有的疫苗尚不能完全有效地预防所有PRRSV流行毒株.尤其是近年来,疫苗免疫压力和猪场种猪的引种导致我国出现PRRSV新亚型,如近年来在我国出现的NADC30 like毒株,所以开展该病毒遗传进化研究,对猪场采取何种疫苗进行蓝耳病的防治具有重要意义.赵津等[10]研究了猪病毒病PCV2,PEDV,TGEV,GAR的复合PCR方法建立及应用;陈燕君等[11]研究了猪诺如病毒巢式RT-PCR检测方法的建立与应用;Chen等[12]指出猪繁殖与呼吸综合征病毒结构基因ORF5对该病毒分子流行病学、致病机理、疫苗研制和鉴别诊断具有重要的意义.因此,本研究拟通过采集2015-2016年本地区疑似感染PRRSV(病料、血清等)的猪群,用RT-PCR扩增其ORF5基因,并进行克隆、序列测定和遗传进化分析,了解2015-2016川南感染PRRSV猪群中ORF5基因的遗传变异情况,对该地区疫苗的选用及PRRSV防控提供依据.
全文HTML
-
无菌采集疑似感染猪繁殖与呼吸综合征高热病猪血清样品、高热病死猪肺脏、淋巴结、脾脏等病料,使用样品碾磨器将所采取组织研磨,血清和所研磨组织液2 000 r/min离心3 min,取上清,冻于-80 ℃保存备用.所处理的样品使用世纪元亨猪繁殖与呼吸综合征RT-PCR检测试剂盒进行检测,操作步骤按照试剂盒说明书进行.
-
采用Omega公司RNA提取试剂盒PRRSV阳性样品全RNA,使用大连宝生物反转录试剂盒primeScript (Takara,大连)对所提取RNA进行cDNA合成.使用ORF5引物序列扩增ORF5基因[11],其PCR扩增程序为94 ℃ 4 min;94 ℃ 40 s,53 ℃ 45 s,72 ℃ 1 min,共29个循环;72 ℃ 10 min;4 ℃保存. PCR产物经1.0%琼脂糖凝胶电泳观察结果(BIO-RAD).将胶块中目的DNA,采用Tiangen胶回收试剂盒回收目的基因,将所回收目的基因与大连宝生物pMD-19T载体进行TA克隆,转化DH5α,挑取阳性克隆鉴定,阳性克隆子送上海生工测序.并将所测定序列与Genbank进行比对,去除猪场所使用疫苗JXA1,HuN4,TJM-F92和CH-1a毒株一致的序列后,所剩序列为猪场感染病毒毒株基因.
-
将所测ORF5序列与在Genbank中所下载的国内外28条ORF5代表序列(表 1),如VR2332,CH-1a,JXA1,Hun4等代表毒株使用Lasergene软件Megalign进行核酸序列相似性和氨基酸序列相似性分析,使用Mega 7.0软件构建其遗传进化树.
1.1. 病料采集、处理及PRRSV检测
1.2. PRRSV阳性样品ORF5基因扩增、克隆及序列测定
1.3. ORF5基因序列分析
-
从所采集的85份标本中,通过使用猪繁殖与呼吸综合征病毒检测试剂盒检测出29份PRRSV PCR阳性,使用ORF5基因特异性引物扩增29份阳性样品的ORF5基因片段,将所获得的29个基因片段送上海生工测序,得到29个ORF5序列,除掉14个与疫苗毒株JXA1,TJ,HuN4核酸序列100%相同外,获得了15个猪繁殖与呼吸综合征病毒ORF5基因序列,将其命名为SC1501-SC1511,SC1601-SC1604.
-
将获得的15个PRRSV分离株的ORF5基因序列与从Genbank中下载的ORF5基因序列比较发现,15个ORF5基因序列均为北美型,15个ORF5遗传距离与序列相似性分别为0.0~0.25和81.1%~100% (表 2),与北美代表毒株ATCC-VR2332毒株的核酸序列相似性和氨基酸相似性分别为83.4%~89.1%和82.1%~88.1 %,与我国2007年流行较广的HP-PRRSV代表毒株JXA1-like核酸序列相似性和氨基酸相似性分别为83.4%~99.3%和83.1%~99.5%,与近年来从北美流传来的NACD30-like毒株核酸序列相似性和氨基酸相似性分别为81.9%~95.7%和80.6%~94.5%,与我国东部新出现的JX1411核酸序列相似性和氨基酸相似性分别为81.4%~92.9%和81.6%~92.5%(表 3).
-
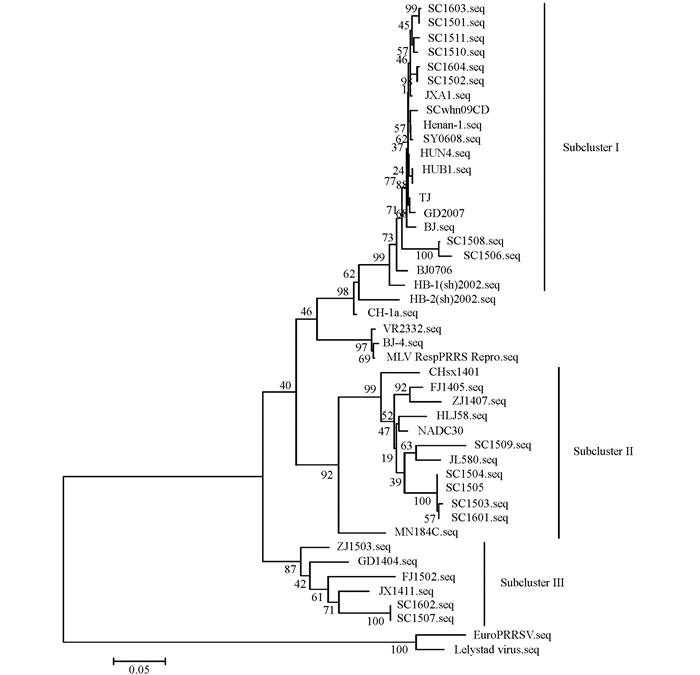
遗传进化分析发现,所测定的15个ORF5基因序列与Genbank中下载的序列形成3个亚群(图 1),SC1501,SC1502,SC1510,SC1511,SC1603,SC1604,SC1506,SC1508等8个ORF5序列与HP-PRRSV类型如JXA1,Hun4,SCwhn09CD等毒株ORF5序列形成1个亚群(subcluster Ⅰ),SC1503,SC1504,SC1505,SC1601,SC1509等5个序列与NADC30,HLJ58,JL580,ZJ1407,FJ1405,HN1502和CHsx1401形成1个亚群(subcluster Ⅱ);SC1507和SC1602基因序列与ZJ1503,FJ1502,JX1411,GD1404和HN1506形成新的1个亚群subcluster Ⅲ.
2.1. 川南地区猪繁殖与呼吸综合征阳性结果
2.2. 川南地区猪繁殖与呼吸综合征ORF5序列同源性分析
2.3. 川南地区猪繁殖与呼吸综合征ORF序列
-
规模化猪场持续性感染猪繁殖与呼吸综合征病毒给养猪业带来严重的经济损失[6, 13].尽管各国都采取了多种防疫措施如疫苗计划免疫与多种综合防疫,但该病仍不断发生,其主要原因是PRRSV遗传变异频率高,对所免疫疫苗出现免疫逃避机制,而现有疫苗的研究速度不能赶上病毒的变异.此外,由于引种等带来的多基因型与本地区现有基因型发生重组而出现多种新的基因亚型,如目前我国出现北美基因型的如CH-1a,JXA1及近年来在我国报道的NADC30毒株[7].本研究通过对获得的15个猪繁殖与呼吸综合征ORF5基因序列进行分析,得出其均属于美洲型毒株,其中8株PRRSV分离株与HP-PRRSV代表株JXA1和HUN4具有较高同源性,表明本地区HP-PRRSV仍然是主要毒株类型.此外,SC1503,SC1504,SC1505,SC1601,SC1509等5个序列与NADC30,HLJ58,JL580,ZJ1407,FJ1405,HN1502和CHsx1401形成1个亚群. NADC30毒株2010年在美国被首次发现[14],近3年我国不断有该类型毒株的报道,如HLJ58,JL580,ZJ1407,FJ1405,HN1502和CHsx1401,这种情况可能与我国从北美进口种猪及猪肉等因素有关,要加强进口种猪及肉制品的检验检疫工作.但是,这些NADC30-like毒株与NADC30序列存在变异现象,容易形成新的毒株类型[7-8].前期研究表明,通过进口种猪及肉制品,NADC30毒株可能已经与我国的HP-PRRSV like毒株发生基因重组,形成了新的基因型猪繁殖与呼吸综合征病毒如ZJ1503,FJ1502,JX1411等,本地区也发现了此类型的PRRSV毒株,如SC1507与SC1602等[9].
川南地区所流行的猪繁殖与呼吸综合征病毒目前仍然以HP-PRRSV毒株为主,存在可能通过引种及进口猪肉制品引进的NADC30类型的PRRSV毒株,出现了可能由于以上2种类型毒株通过重组形成的新类型PRRSV的ZJ1503-like毒株.笔者推测,通过严格控制引种及肉制品引进,减少新毒株与本地区毒株发生基因重组出现复杂的PRRSV基因型,对PRRSV的防控具有重要意义.



 下载:
下载: