-
植物内生菌(Endophyte)指在生活史的部分阶段或全部阶段生活在植物的根、茎或者叶片等组织内,但不会引起病害症状的一类微生物[1].常见的植物内生菌包括真菌、放线菌和细菌等,它们是植物微生物态系统的重要组成部分,几乎每一种植物体内都能发现内生菌,这些内生菌通过产生对植物具有保护或者促进作用的代谢产物促进植物生长并最终有利于自身的生长[2-3].植物内生菌还能产生各种具有生物活性的次级代谢产物,例如紫杉醇等[4].部分植物内生菌产生的次级代谢产物被发现在医药工业与病虫害防控上具有潜在的应用价值,使得植物内生菌成为了近年来研究的热点[5].
许多研究已经证实昆虫的共生肠道微生物对宿主的生长产生着有利的作用,例如通过调节微生物的代谢,提升消化食物的效率使得昆虫能够从食物中获得更多的营养,以及保护宿主抵抗病原菌的感染等[6-8].昆虫肠道微生物也与昆虫的农药抗性相关,小菜蛾肠道微生物中的肠球菌可以提升宿主的农药耐药性,而另一种微生物沙雷氏菌则会降低宿主的农药抗性[9].许多昆虫以采食植物组织为生,在这个过程中必然会摄入大量植物内生菌进入昆虫肠道中,而这些植物内生菌与昆虫肠道微生物组的关系较为复杂,人们对此还缺乏深入的了解.部分植物内生真菌可以赋予宿主一定的抗虫能力,例如定植在玉米中的白僵菌可以减少欧洲玉米螟的危害[10].韩国学者的一项研究表明昆虫肠道微生物群的主要影响因素包括栖息地、食物、发育阶段和昆虫的种类等[11].中国学者发现在采食桑叶的不同昆虫(家蚕、桑剑纹夜蛾和桑螟)肠道中,微生物的多样性存在较大的差异[12].而另一项研究则发现,5种采食苏铁的昆虫具有相似的肠道核心微生物组[13].
草地贪夜蛾是一种危害巨大的迁飞性农林害虫,可采食玉米、水稻、高粱和甘蔗等重要农作物.该害虫自2019年1月入侵中国以来,已经蔓延至全国21个省份,严重威胁我国粮食生产.本课题组前期对草地贪夜蛾肠道优势细菌进行了初步的分离鉴定工作,并对取食不同植物的草地贪夜蛾肠道微生物群也进行了比较,对研究草地贪夜蛾核心肠道微生物提供了重要的理论支持[14-15].鉴于草地贪夜蛾的肠道微生物中必然有一些源自于其取食植物的内生菌,因此研究其食材中的内生菌就成为了进一步解析草地贪夜蛾肠道微生物群的基础.基于此,本研究以采集自重庆巫山地区爆发草地贪夜蛾虫害的玉米田内的玉米嫩叶为样本,通过传统培养法以及16S rDNA测序对其中的植物内生细菌进行了初步分离鉴定,以期为后续的研究奠定基础.
全文HTML
-
草地贪夜蛾幼虫和玉米茎叶由巫山县植保站在野外玉米地中采集后送至西南大学.
-
马铃薯琼脂培养基(PDA):马铃薯200.0 g,葡萄糖20.0 g,琼脂15.0 g,用蒸馏水定容至1 000 mL.
LB培养基:胰蛋白胨10 g,酵母提取物5 g,NaCl 10 g,用蒸馏水定容至1 000 mL.
牛肉膏蛋白胨培养基(NA):牛肉膏5.0 g,蛋白胨10.0 g,NaCl 5 g,用蒸馏水定容至1 000 mL.
-
SDS,CATB,NaCl,Tris,EDTA、蛋白酶K、蔗糖和溶菌酶均购自生工生物工程(上海)有限公司;苯酚、氯仿和异戊醇购自重庆川东化工(集团)有限公司;PCR扩增引物27F:5′-AGAGTTTGATCCTG GCTCAG-3′;1492R:5′-GGTTACCTTGTTACGACTT-3′,由生工生物工程(上海)股份有限公司合成;1×Taq MasterMix(purple)购自北京博迈德基因技术有限公司.
-
在超净台中,将玉米嫩叶以75%的乙醇进行表面消毒.表面消毒完成后,取出适量叶片组织进行研磨匀浆后作为原液备用.
-
取100 μL制备的匀浆液于新的1.5 mL离心管中,加入900 μL灭菌PBS溶液配制成10-1组,随后递次稀释为10-2至10-6备用.分别取10-2组至10-6组溶液100 μL涂布于不同的固体培养基上,37 ℃培养箱中培养72 h,每24 h观察一次并随机挑选单菌落,在分离培养基上进行划线获得纯培养.
-
1) 取适量菌液离心收集菌体后,以500 μL TE25S缓冲液(25 mmol/L Tris-HCl,25 mmol/L EDTA,0.3 mol/L蔗糖)重悬.
2) 加入10 μL溶菌酶溶液(100 g/L),在37 ℃温度下静置30~60 min.
3) 加入5 μL蛋白酶K溶液(20 g/L)和30 μL 10% SDS,55 ℃水浴60 min.
4) 加入100 μL 5 mol/L NaCl,上下颠倒混匀.
5) 加入65 μL CTAB/NaCl(0.7 mol/L NaCl,10% CTAB)混合液,混合均匀后在55 ℃温度下静置10 min.
6) 取出冷却至室温,加入400 μL酚/氯仿/异戊醇混合液(各组分体积比为25:24:1),震荡混匀5~10 min.
7) 以12 000 r/min的转速离心10 min.
8) 将上清液转移至新的1.5 mL离心管中,重复步骤6-7.
9) 将上清液转移至新的1.5 mL离心管中,加入等体积异丙醇,上下颠倒混匀.
10) 以12 000 r/min的转速离心10 min,弃去上清液,加入70%乙醇后颠倒混匀,再以12 000 r/min的转速离心10 min,重复该操作1次.
11) 弃去上清液,打开离心管盖静置5~10 min,晾干基因组DNA,加入50~100 μL ddH2O溶解DNA.
-
1) 用细菌通用引物27F和1492R对上述模板进行PCR扩增细菌16S rDNA序列.反应体系(50 μL)如下:模板5 μL,引物27F/1492R各1 μL,1×Taq MasterMix(purple) 43 μL.反应程序为:96 ℃预变性10 min;96 ℃变性30 s,55 ℃退火30 s,72 ℃延伸1 min 30 s,30个循环;72 ℃延伸10min.将扩增产物送至生工生物工程(上海)股份有限公司测序.
2) 将获得的16S rDNA基因序列在GenBank(http://blast.ncbi.nlm.nih.gov/Blast.cgi)与数据库已登录基因序列进行同源比对,下载GenBank中同源性较高序列,利用MEGA 6.0软件[16],使用N-J法进行1 000次步长计算,构建系统发育树.同时将获得的16S rDNA基因序列在RDP数据的Classifer程序(https://rdp.cme.msu.edu/classifier/classifier.jsp)进行比对,获得同源性较高序列的相关信息.
1.1. 样本材料
1.2. 实验用的培养基
1.3. 主要试剂
1.4. 玉米叶的初步处理
1.5. 植物内生细菌的分离
1.6. 分离菌株的初步鉴定
1.6.1. 细菌总DNA的提取
1.6.2. 菌株的分子生物学鉴定
-
采用PDA培养基、LBG培养基和NA培养基分离获得了22个分离株.通过细菌通用引物27F/1492R对22个分离株进行PCR扩增测序获得其16S rDNA序列.将所有测序结果利用Blastclust进行同源性聚类分析,同源性阈值97%,共获得11个分类单元——OTU 1-11(表 1).
将测序结果分别在GenBank和RDP两个数据库比对获得相关信息.比对结果显示,本次从巫山地区的玉米叶中分离的22个分离株中,WSma3,WSma4,WSma5,WSma11和WSma12属于不动杆菌属(Acinetobacter),WSma8,WSma9和WSma20属于肠球菌属(Enterococcus),WSma1和WSma2属于克雷伯氏菌属(Klebsiella),WSma6和WSma7属于芽孢杆菌属(Bacillus),WSma10,WSma14和WSma16属于微杆菌属(Microbacterium),WSma18和WSma19属于鞘氨醇杆菌属(Sphingobacterium),WSma21和WSma22属于金黄杆菌属(Chryseobacterium),WSma17属于无色杆菌属(Achromobacter),WSma13属于丛毛单胞菌属(Comamonas),WSma15属于寡养单胞菌属(Stenotrophomonas);在所有分离株中,不动杆菌属的相对丰度最高(表 1).
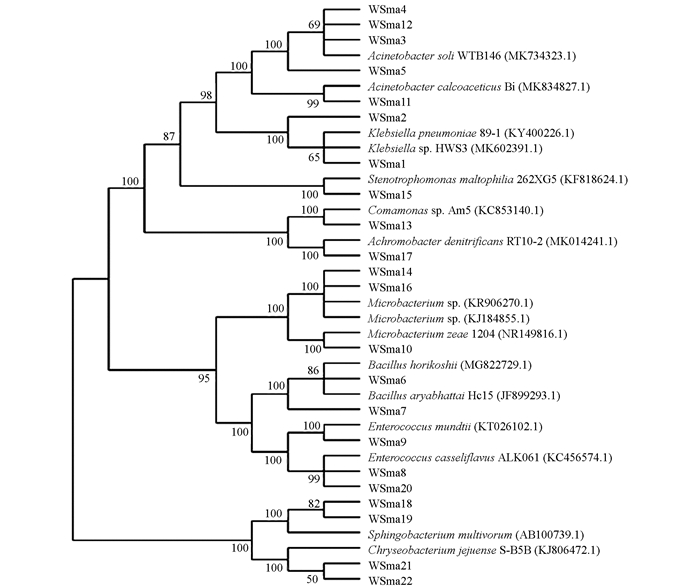
为进一步了解巫山地区玉米叶内生细菌的遗传多样性,对22个植物内生细菌的分离株进行了系统发育分析.对分离株16S rDNA序列进行BLAST比对分析,采用MEGA 6.0软件N-J法构建系统发育树(图 1).
由图 1可见,22个分离株在进化发育树上分属10个属,分别为不动杆菌属(Acinetobacter)、肠球菌属(Enterococcus)、克雷伯氏菌属(Klebsiella)、芽孢杆菌属(Bacillus)、微杆菌属(Microbacterium)、鞘氨醇杆菌属(Sphingobacterium)、金黄杆菌属(Chryseobacterium)、无色杆菌属(Achromobacter)、丛毛单胞菌属(Comamonas)和寡养单胞菌属(Stenotrophomonas).其中5株属于不动杆菌菌属(Klebsiella),占整体的27%. WSma8,WSma9和WSma20都属于肠球菌属,但聚类为OTU4的WSma8和WSma20与聚类到OTU8的WSma9在进化关系上存在一定的差异.
-
本研究通过纯培养的方法,分离培养并初步鉴定了巫山地区爆发草地贪夜蛾虫害的玉米地中玉米叶的内生细菌,共鉴定到了10个属的细菌,分别为不动杆菌属(Acinetobacter)、肠球菌属(Enterococcus)、克雷伯氏菌属(Klebsiella)、芽孢杆菌属(Bacillus)、微杆菌属(Microbacterium)、鞘氨醇杆菌属(Sphingobacterium)、金黄杆菌属(Chryseobacterium)、无色杆菌属(Achromobacter)、丛毛单胞菌属(Comamonas)和寡养单胞菌属(Stenotrophomonas),这些属的微生物均有报道证明其是常见的玉米内生菌[17].部分内生细菌可以为植物生长提供有利因素,例如芽孢杆菌属微生物可以产生抗真菌物质并促进玉米植株抗性基因表达,提升植物的抗病能力[18];克雷伯氏菌属微生物具有一定的固氮功能,可以为植物提供一定程度的营养[19].本课题组研究发现巫山玉米地中采集到的草地贪夜蛾肠道中存在多种微生物,包括克雷伯氏菌属(Klebsiella)、不动杆菌属(Acinetobacter)、肠杆菌属(Enterobacter)、气单胞菌属(Aeromonas)、短波单胞菌属(Brevundimonas)、金黄杆菌属(Chryseobacterium)和肠球菌属(Enterococcus)[14-15, 20],后续研究还进一步鉴定到了鞘氨醇杆菌属(Sphingobacterium)和类芽孢杆菌属(Paenibacillus)(数据未发表).通过比较可以发现在草地贪夜蛾肠道中分离纯化得到的微生物很多也同样可以在玉米叶中分离得到,例如鞘氨醇杆菌属(Sphingobacterium)、不动杆菌属(Acinetobacter)、克雷伯氏菌属(Klebsiella)和金黄杆菌属(Chryseobacterium)等,表明草地贪夜蛾的肠道微生物有很多都可能源自其取食的植物内生菌.不过也有一些微生物在两者之间存在差异,例如我们仅在玉米叶中分离得到芽孢杆菌属(Bacillus)、寡养单胞菌属(Stenotrophomonas)、丛毛单胞菌属(Comamonas)、微杆菌属(Microbacterium)和无色杆菌属(Achromobacter)细菌,并未在草地贪夜蛾的肠道微生物群中发现这些属的细菌.推测这些微生物可能并不适应草地贪夜蛾肠道环境,因此我们在草地贪夜蛾肠道微生物群中未能分离到相应的微生物.而相对地,在玉米叶内生菌中未发现草地贪夜蛾肠道中常见的类芽孢杆菌属(Paenibacillus)、肠杆菌属(Enterobacter)、气单胞菌属(Aeromonas)、短波单胞菌属(Brevundimonas)和假单胞菌属(Pseudomonas)微生物,表明这些微生物不是源于植物内生菌,而是为草地贪夜蛾所独有,可能源自环境中摄入或者孵化后取食卵壳.
类芽孢杆菌属(Paenibacillus)是一种常见于玉米根部以及周围土壤环境中的植物内生菌[21],本研究从草地贪夜蛾肠道微生物群中分离到了类芽孢杆菌属细菌,而在玉米叶片中则未分离到该菌.最近有学者认为取食叶片昆虫的肠道微生物组更多地受到土壤微生物组的影响[22],本研究结果从一定程度上支持了该研究的论点.有研究表明不动杆菌属(Acinetobacter)的微生物可以降解杀虫剂进而改变宿主的农药抗药性[23],在玉米叶与草地贪夜蛾中均分离到了不动杆菌属的微生物,在后续的研究中我们将分析玉米叶与草地贪夜蛾肠道中分离得到的不动杆菌属微生物是否也具有相应的能力.
前期研究发现以高粱为食的草地贪夜蛾肠道细菌与以玉米为食的草地贪夜蛾肠道细菌中均存在不动杆菌属(Acinetobacter)和假单胞菌属(Pseudomonas)的微生物[15],说明这些微生物可能对草地贪夜蛾具有重要的意义,而本研究证实玉米叶中假单胞菌属微生物相对数量较少,进一步支持了该结论.前期研究在取食高粱叶的草地贪夜蛾肠道中未分离出克雷伯氏菌属(Klebsiella),而在取食玉米的草地贪夜蛾肠道中分离到了克雷伯氏菌,本研究发现玉米叶中亦存在克雷伯氏菌,这表明该差异可能源于取食植物的不同,但该观点需要分析高粱叶中微生物组成来进一步验证.
综上,我们报道了重庆巫山地区暴发草地贪夜蛾玉米地中玉米叶的内生细菌分离株,发现其组成与取食该玉米叶的草地贪夜蛾肠道微生物群存在一定的相似性与差异性,这有助于进一步阐明草地贪夜蛾的核心微生物群.至于这些植物内生菌在草地贪夜蛾肠道的活跃程度如何,它们是否会影响着宿主的抗性、生长发育和迁飞等问题,仍有待进一步研究.



 下载:
下载: