-
开放科学(资源服务)标志码(OSID):

-
多聚半乳糖醛酸酶(Polygalacturonase,PG)是一种参与多种植物发育过程的果胶消化酶. 果胶是一种杂多糖,它是双子叶植物体细胞初生细胞壁的主要成分,同时也是花粉内壁和花粉管壁的主要成分,因此,参与果胶代谢相关酶和蛋白对植物的生长和生殖发育至关重要[1]. PG基因属于植物中的大基因家族,在拟南芥[2]、杨树[3]、苹果[4]、葡萄[5]和玉米[6]等物种中均进行了PG家族基因的鉴定和表达模式研究. PG基因参与植物发育的不同阶段,如种子萌发,器官脱落,荚和花药开裂,花粉粒成熟,花粉管生长,和木质部细胞形成[7]. 在水稻中的超表达PG基因降低了果胶组分和细胞粘着力,同时,还增加了对非生物逆境的敏感性[8];而在苹果中PG基因的超表达导致叶片形态发育不正常,果实脱落等表型[9]. PG基因的表达水平在不同器官和组织中显著不同[10],超过50%的拟南芥PG基因在花卉组织中高度表达[11]. 拟南芥中的PGX1参与细胞的伸长、膨胀和花的生长和发育;PGX2能够促进拟南芥子叶下胚轴的伸长、叶的扩张、茎的木质化等;QRT2、ADPG1和ADPG2对拟南芥生殖发育过程中花粉粒等不同的细胞分裂具有很重要的作用[12-14].
目前,栽培甜荞均为自交不亲和的两型花作物,包括长柱花和短柱花,长柱花(L-morph Flower)也称“Pin”型花,具有长花柱和短雄蕊;短柱花(S-morph Flower)也称“Thrum”型花,具有短花柱和长雄蕊. 这种花粉的互位方式减少了花粉的浪费,促进了异花授粉,提供了花粉的适应性[15],同时,也极易受到环境影响,开花期出现阴雨天则会导致结实率降低而影响产量. 国内外多个研究团队已利用自交可育野生荞麦Fagopyrum homotropicum[16],通过种间杂交的方式(Fagopyrum esculentum和F.homotropicum)培育出自交可育的等柱花甜荞,即花柱与雄蕊等长,且自交可育. 依据实验室前期对甜荞长、短和等柱花雌雄蕊转录组数据,获得一个PG基因编码序列,命名为FePG1,本研究拟对FePG1基因进行基因克隆,系统进化分析,亚细胞定位及表达模式分析,以期为今后验证甜荞PG基因家族成员调控甜荞花柱异长及作用机理提供理论参考.
全文HTML
-
以花柱异长的自交不亲和甜荞品种乌克兰大粒荞和酉荞2号,花柱等长的自交亲和甜荞品种甜自21和贵甜2号为试验材料. 种植于西南大学科研基地,于盛花期采集开花当日长势一致的花朵收集其雌雄蕊,置于-80 ℃冰箱中保存备用. 此外,于盛花期取乌克兰大粒荞短柱花的根、茎、叶、花和幼嫩籽粒,置于-80 ℃冰箱中保存备用.
利用Biospin多糖多酚植物总RNA提取试剂盒(博日,杭州)提取RNA,再经琼脂糖电泳检测RNA降解和污染情况. 用RNA Nano 6000 Assay Kit of the Agilent生物分析仪2100 system (Agilent Technologies,CA,USA)检测RNA完整性,用NanoPhotometer®分光光度计(IMPLEN,CA,USA)检测RNA的纯度,用Qubit® RNA Assay Kit in Qubit® 2.0荧光计(Life Technologies,CA,USA)检测RNA的浓度. 利用PrimeScriptTMRT reagent Kit with gDNA Eraser(Perfect Real Time)(TaKaRa,北京)试剂盒进行cDNA的合成,于-80 ℃冰箱中保存备用.
-
实验室前期已利用乌克兰大粒荞长柱花、短柱花和甜自21等柱花雌雄蕊总RNA进行了转录组测序,分析测序结果发现FePG1基因在乌克兰大粒荞短柱花中高表达、在等柱花中不表达. 本研究以乌克兰大粒荞短柱花雌雄蕊cDNA为模板,引物序列FePG1-F(5'-ATGGCCACGTTTTGGATTTGTATG-3')和FePG1-R(5'-TAGCAGTGAACAGGAGGAAGC-3'),利用PrimeSTAR Max Premix(2X)聚合酶(TaKaRa,北京)进行PCR扩增. 扩增产物用1%的琼脂糖凝胶电泳进行检测,用SanPrep柱式DNA凝胶回收试剂盒(Sangon Biotech,上海)对扩增的目的片段进行回收. 将目的基因导入到pMD19-T(TaKaRa,北京)中,经蓝白斑筛选,挑取白色单菌落到100 μg/mL的LB(Luria-Bertani)液体培养基中培养后,送至北京华大基因进行测序.
-
将测序正确的甜荞FePG1基因序列翻译成氨基酸,通过ExPASy-prosite (http//www.expasy.org)分析甜荞FePG1基因的CDS序列保守结构域. 编码蛋白理化性质分析和蛋白亲水性预测利用在线工具ProtParam(https://web.expasy.org/protparam/)进行分析和预测;编码蛋白的跨膜结构分析通过TMpred工具(https://embnet.vital-it.ch/software/TMPRED_form.html)进行分析;编码蛋白信号肽预测利用在线工具SignalP(http://www.cbs.dtu.dk/services/SignalP/)进行预测. FePG1基因的蛋白序列在NCBI(National Center for Biotechnology Information)数据库中执行BlastP搜索(https://blast.ncbi.nlm.nih.gov/Blast.cgi). 选取来源于30个不同被子植物的PG同源蛋白,采用MEGA 7.0软件的邻接法(NJ)构建蛋白序列的系统发育树.
-
分别以两个自交不亲和甜荞品种乌克兰大粒荞和酉荞2号长柱花、短柱花雌雄蕊cDNA及两个自交亲和甜荞品种甜自21和贵甜2号等柱花雌雄蕊cDNA为模板,用Primer Primer 5.0软件设计基因特异性引物FePG1-F(5'-CCGGAATTCATGGCCACGTTTTGGATTTG-3')和FePG1-R(5'-TGCTCTAGATTAGCAGTGAACAGGAGGAAGC-3'). 使用TB GreenTM Premix Ex TaqTM Ⅱ (Tli RNaseH Plus)试剂盒(TaKaRa,北京)检测甜荞FePG1基因在不同花柱类型中的表达差异. 以乌克兰大粒荞短柱花的根、茎、叶、花和幼嫩籽粒cDNA为模板(PCR反应体系和循环参数同上),检测甜荞FePG1基因的组织表达情况.
-
将本实验室保存的烟草种子播种至装满基质土的花盆(15 cm×15 cm×12 cm)中,花盆置于光照培养箱中培养,培养条件为光周期16/8(光照/黑暗) h、温度25 ℃、湿度60%,待培养4周后备用. 根据目的基因FePG1的序列设计带有StuI酶切位点的引物FePG1-F(5'-AAAAGGCCTATGGCCACGTTTTGGATTTG-3')和FePG1-R(5'-TTTTCCGGATTAGCAGTGAACAGGAGGAAGC-3'),扩增FePG1基因的ORF(开放阅读框)区域,扩增产物经切胶回收. 用限制性内切酶StuI对pH=7 LIC5.0-TET2rc-N-eGFP进行单酶切. 酶切产物用1%琼脂糖凝胶电泳,回收纯化目的条带,用T4连接酶将扩增产物和酶切产物经12 h连接,而后转化大肠杆菌DH5α,12 h培养,挑取阳性克隆进行菌液PCR验证,验证成功的单克隆子扩大培养,提取重组质粒并进行酶切验证. 依据pH=7 LIC5.0-TET2rc-N-eGFP的StuI酶切骨架设计引物F(5'- ACGTCTATATCATGGCCGACA-3')和R(5'-GTGACTCCCTTAATTCTCATGTATGATA-3'),将得到重组质粒pH=7 LIC5.0-N-eGFP-FePG1测序验证. 将重组质粒pH=7 LIC5.0-N-eGFP-FePG1和对照质粒pH=7 LIC5.0-TET2rc-N-eGFP分别转化农杆菌感受态GV3101. 将菌液进行PCR检测后再注射到烟草叶片背面,经过常温黑暗处理12 h后观察叶片信号. 切取侵染3 d后的烟草叶片进行制片,置于激光共聚焦显微镜下观察荧光在烟草下表皮细胞内的分布情况.
1.1. 供试材料
1.2. 甜荞FePG1基因克隆
1.3. 甜荞FePG1基因生物信息学分析
1.4. 甜荞FePG1基因的表达分析
1.5. 甜荞FePG1基因的亚细胞定位
-
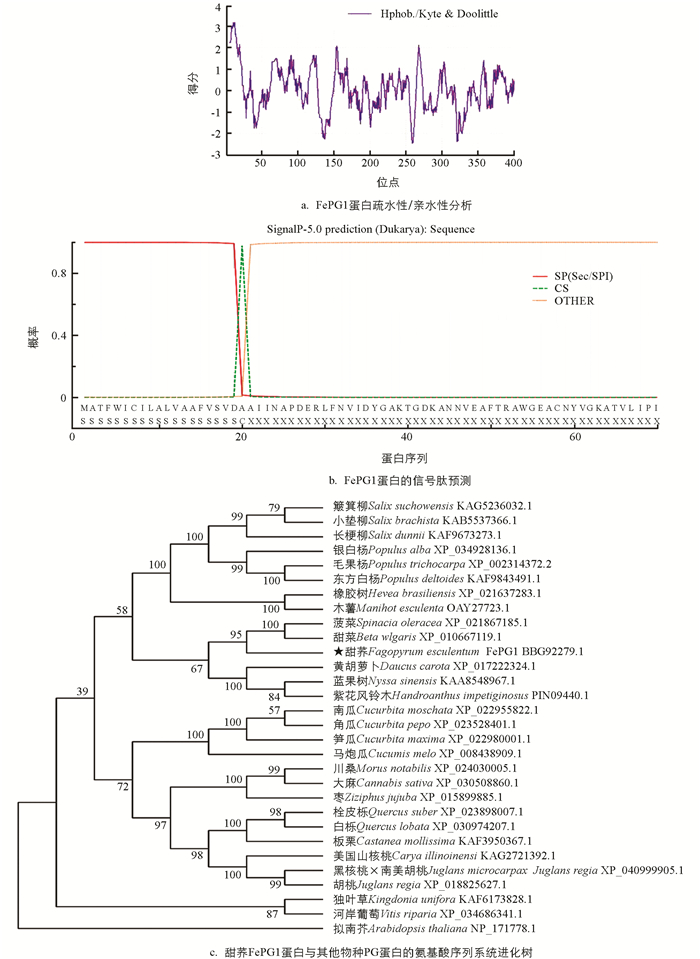
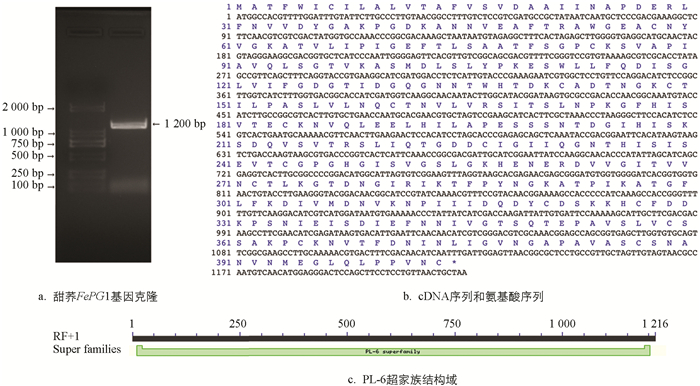
利用前期的转录组数据,以花柱异长的自交不亲和甜荞品种乌克兰大粒荞短柱花雌雄蕊cDNA为模板,通过PCR扩增得到FePG1基因(图 1a). 分析FePG1基因的CDS片段,全长1 215 bp,编码404个氨基酸(图 1b). 相对分子质量为4.3×104,化学分子式为C1895H3018N508O596S18,等电点为5.19.其中包括30个亲水性氨基酸(Arg+Lys)、42个酸性氨基酸(Asp+Glu)、39个碱性氨基酸(Arg+Lys+His)和132个疏水性氨基酸(Ala,Lle,Leu,Met,Val). 使用NCBI中Conserved Domains对FePG1蛋白序列进行功能结构域分析,结果表明该蛋白含有一个典型PL-6(多糖裂解酶,大多与果胶的形成及功能关系密切)超家族结构域(图 1c),占该基因序列的90%以上.
-
利用ProtParam预测得到FePG1蛋白亲水性平均系数为+0.058,脂肪族氨基酸指数为95.30,再利用ProtScal进一步分析疏水性氨基酸多于亲水性氨基酸,疏水性最大值为3.189(第10位和第12位的氨基酸),最小值为-2.467(第259位的氨基酸)(图 2a),推测FePG1基因编码的蛋白为疏水性蛋白.
通过信号肽预测在线工具SignalP分析得出,FePG1蛋白为信号肽的可能性为0.999,其他类型仅0.001(图 2b). 因此,该蛋白存在信号肽,属于分泌性蛋白.
系统发育分析表明(图 2c),甜荞FePG1蛋白与石竹目藜科作物甜菜、菠菜同源蛋白的亲缘关系较近,其次为大戟科的橡胶树、木薯,以及杨柳科的杨树、柳树等;与拟南芥亲缘关系较远.
-
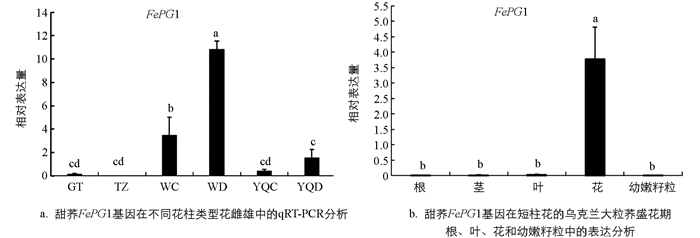
在课题组前期转录组数据基础上,为了验证FePG1基因在甜荞中的表达情况,本研究分别提取了盛花期乌克兰大粒荞长柱和短柱花雌雄蕊、甜自21等柱花雌雄蕊、酉荞2号长柱和短柱花雌雄蕊、贵甜2号等柱花雌雄蕊进行验证. 通过qRT-PCR技术分析,结果表明FePG1基因在自交亲和甜荞品种贵甜2号和甜自21的雌雄蕊中几乎不表达,在自交不亲和甜荞乌克兰大粒荞和酉荞2号中高表达,且在短柱花雌雄蕊表达量均显著高于长柱花雌雄蕊(图 3a).
为了进一步分析FePG1基因的组织表达特异性,提取盛花期乌克兰大粒荞的根、茎、叶、花和幼嫩籽粒通过qRT-PCR进行表达验证,结果表明FePG1基因只在花中高表达,在根、茎、叶和幼嫩籽粒中基本不表达(图 3b),在甜荞花中特异性表达.
-
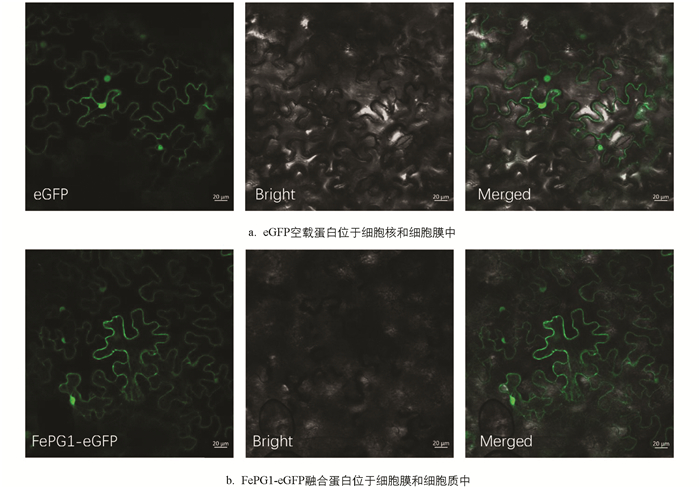
FePG1基因注释为转录因子,为了在亚细胞水平上研究FePG1基因的表达情况,利用注射法侵染烟草下表皮观察FePG1-eGFP融合蛋白和eGFP空载蛋白. 以eGFP空载蛋白为对照,由于其在细胞中的定位没有特异性,所以在细胞膜和细胞核中观察到荧光信号(图 4a). 在烟草叶片细胞中检测到FePG1-eGFP分布较多,主要集中于细胞膜和细胞质中(图 4b).
2.1. FePG1基因克隆
2.2. 甜荞FePG1蛋白序列及分子系统发育分析
2.3. FePG1基因的表达验证及表达模式分析
2.4. FePG1基因的亚细胞定位分析
-
甜荞属于蓼科荞麦属作物. 前人对荞麦属作物甜荞、苦荞和金荞麦籽粒转录组测序结果表明,与荞麦属同源基因最多的物种是甜荞(23%)、葡萄(11.3%)[17]. 对甜荞花序[18]、籽粒[19]转录组测序结果表明甜荞与葡萄、碧桃、蓖麻、毛果杨亲缘关系较近. 本研究从甜荞雌雄蕊中克隆到的FePG1蛋白,经系统发育分析,与石竹目藜科作物甜菜、菠菜,以及大戟科和杨柳科同源蛋白的亲缘关系较近,与前人研究结果一致.
多聚半乳糖醛酸酶在细胞分离的生命周期过程中起着重要作用,对拟南芥细胞壁修饰、脱落和开裂均有重要影响[12]. 50%以上拟南芥的PG基因在花组织中高表达[11]. 本文研究基于前期转录组测序克隆了一个FePG1基因,利用两个自交亲和甜荞和两个自交不亲和甜荞为材料,qRT-PCR实验表明FePG1基因在自交亲和甜荞雌雄蕊中不表达,在自交不亲和甜荞中表达,且其表达量在短柱花雌雄蕊中显著高于长柱花,表明FePG1基因可能与甜荞自交不亲和有关,且与花柱异长发育有关.
多聚半乳糖醛酸酶是GH28家族的果胶降解糖苷水解酶的重要成员之一,GH28亚细胞定位显示存在针对分泌途径的信号序列[7]. 本研究通过亚细胞定位证明FePG1蛋白为存在于细胞膜和细胞质的跨膜蛋白,与本实验FePG1蛋白的跨膜结构预测结果相吻合. 据此,我们推断FePG1基因编码是一种存在于细胞膜和细胞质中的跨膜蛋白,但是否与分泌途径有关系需进一步探究.



 下载:
下载: