-
开放科学(资源服务)标志码(OSID):

-
组蛋白修饰是最重要的表观遗传修饰标记之一,在控制基因表达和细胞谱系规范中起到关键作用[1-3]. 已有研究表明,组蛋白甲基化对早期胚胎发育和胚胎干细胞多能性的维持具有重要作用[4]. 组蛋白甲基化发生在组蛋白的赖氨酸和精氨酸残基上,作用位点在其侧链.
在N原子上,常见位点有H3K4me3,H3K9me3和H3K27me3[5-6]. 其中,组蛋白H3第9位赖氨酸三甲基化(Histone H3K9 trimethylation,H3K9me3)与维持异染色质稳定和沉默、X染色体失活、印记相关基因抑制及细胞特异性的鉴定有关[7],也被认为是胚胎发育的关键障碍[8].
H3K9me3被证明是小鼠[9-10]、人类[11]和牛[12]重新编程中关键的一个表观遗传调节因子. 在小鼠GV卵母细胞中,H3K9me3的表达与组蛋白甲基转移酶的活性密切相关. BHPF(Bateosi High Pass Filter)处理通过影响细胞骨架结构、线粒体功能、氧化应激、细胞凋亡提高了H3K9me3的表达,从而降低了小鼠卵母细胞的成熟效率和卵母细胞的质量[13]. 大剂量真菌毒素导致小鼠卵母细胞H3K9me3水平升高,这可能是导致卵母细胞发育能力下降的原因之一[14]. 以上研究表明,适当降低H3K9me3的表达有利于卵母细胞体外成熟和早期胚胎发育.
H3K9me3在基因表达调控和卵母细胞生长中起重要作用[15],目前其在体外成熟猪卵母细胞和孤雌胚胎中的表达模式还鲜有报道. 毛壳素作为SUV39亚家族甲基转移酶的抑制剂,能够特异性抑制组蛋白甲基转移酶SUV39H1/2和G9A的活性,通过竞争性结合SUV39H1/2[16],与关键残基发生反应,从而调控H3K9me3水平,以此增强表观遗传重编程效率[17]. 因此,本实验通过在猪卵母细胞体外成熟过程中添加毛壳素,调控卵母细胞H3K9me3及其相关基因的表达. 研究结果有助于了解体外成熟卵母细胞和胚胎中H3K9me3表达的动态变化,并为改善猪卵母细胞成熟质量,提高胚胎发育效率奠定基础.
全文HTML
-
本实验所用卵巢均来源于广西南宁市屠宰场,实验所用胚胎为孤雌激活胚胎.
-
将从屠宰场取得的卵巢置于37 ℃生理盐水中,用生理盐水清洗3遍,并用注射器采集卵巢表面直径约3~10 mm的卵泡,挑选形态完好、胞质均匀,周围带有3层以上致密卵丘细胞的卵丘-卵母胞复合体(cumulus-oocyte complexes,COCs),用洗卵液清净后将30~50个卵母细胞放入用矿物油覆盖的一个胚胎培养液(PM)微滴中,每滴为150 μL培养液,[0~22) h将卵母细胞放入含有激素的培养液中进行培养,之后将卵母细胞移入另一个不含激素的培养液中继续培养[22~44] h. 实验组在对照组的基础上添加了不同浓度的毛壳素.
-
实验前先打开热台、融合仪,并安装激活盘. 用提前平衡的电激活液清洗3遍融合槽,然后把新的电激活液放置于融合槽中. 将卵母细胞在融合液中清洗3遍后放于激活槽中进行激活. 将激活过的卵母细胞移入平衡好的胚胎培养液培养15 min,然后移入35 μL的胚胎培养液微滴中进行培养. 24 h统计卵裂率,48 h统计4-细胞率,7 d统计囊胚率.
-
将收集的猪孤雌囊胚放入4%的多聚甲醛固定液中固定24 h,然后放入10 μg/mL细胞核染料(Hoechst)中,避光染色10 min,然后用磷酸盐缓冲液(PBS)清洗3遍,每次5 min,染好的囊胚移入载玻片的抗淬灭剂中,用凡士林封片,在荧光显微镜下观察并做好统计.
-
收集不同时期的卵母细胞和孤雌囊胚放入4%多聚甲醛中4 ℃固定24 h. 固定之后,室温条件下放入阻断液中阻断3次,每次5 min,然后放入1% Triton X-100中,透化30 min,再用阻断液阻断2次,每次5 min,然后用1%牛血清蛋白溶液(BSA)进行非特异性位点封闭1~2 h,封闭之后用T-BSA-PBS清洗3次,每次5 min,把处理好的样品放入H3K9me3的一抗4 ℃过夜. 第2 d把样品放入培养箱孵育30 min,然后用T-BSA-PBS清洗3遍,每次5 min,清洗好之后把样品放入装有二抗的EP管中,避光室温孵育1.5 h,然后用清洗液清洗3遍,每次5 min,最后用10 μg/mL Hoechst 33342复染10 min,用清洗液清洗两遍放入滴有抗淬灭剂的玻片上,用凡士林封片镜检,记录实验数据.
-
收集不同时期的猪卵母细胞和孤雌囊胚,用PBS+PVA清洗3遍,洗去样品所带的培养液和血清等物质后将样品放入装有细胞裂解液的管中. 微量反转录所有步骤均在冰上操作:裂解,将样品放入聚合酶链式反应(PCR)仪中4 ℃裂解15 min;消化,裂解后每管加入1 μL DNase I和1.3 μL 10×DNA buffer,瞬时离心后放入PCR仪中37 ℃消化40 min;终止,消化完每管加入1 μL EDTA,2 μM Random Primer,1 μL mixture deoxynucleotides (dNTPs),瞬时离心后放入PCR仪中65 ℃10 min;反转,终止后依次加入2 μL DTT,4 μL 5×First-Strand Buffer,0.5 μL RNase Inhibitor(RRI),0.25 μL SuperScriptTMⅡ Reverse Transcriptase (SSⅡ),瞬时离心,进行反转录,其条件是25 ℃ 10 min,42 ℃ 90 min,95 ℃ 10 min,4 ℃停止. 将反转录得到的样品存放于-20 ℃备用.
将微量反转录得到的产物作为模板,进行qRT-PCR检测,观察目的基因的表达情况. 反应体系按说明书进行.
-
本实验结果均使用SPSS 22.0软件进行分析,早期胚胎发育率先通过反正弦变化后再进行分析. 每个实验重复至少3次,每次重复实验中至少随机挑选10个以上卵母细胞进行实验. p<0.05表示差异具有统计学意义.
1.1. 材料和试剂
1.2. 猪卵巢的采集及体外成熟培养
1.3. 猪卵母细胞的孤雌激活
1.4. 囊胚细胞总数的鉴定
1.5. 细胞免疫荧光
1.6. 反转录和qRT-PCR
1.7. 结果统计与分析
-
[0~22) h采用不同浓度毛壳素处理,发现2 nmol/L处理组的卵母细胞第一极体排出率显著高于未处理组、5 nmol/L组和10 nmol/L组;4-细胞率显著高于未处理组、5 nmol/L组和10 nmol/L组;2 nmol/L处理组的囊胚率显著高于未处理组和10 nmol/L组(p<0.05);2 nmol/L处理组的囊胚细胞总数显著高于5 nmol/L组和10 nmol/L组(p<0.05),与未处理组之间差异不具有统计学意义(p>0.05) (表 1).
-
[22~44] h采用不同浓度毛壳素处理时发现,与未处理组相比,5 nmol/L毛壳素处理组的第一极体排出率显著提高(87.37% VS 71.91%,p<0.05),各处理组之间的卵裂率、4-细胞率、囊胚率和囊胚细胞总数差异不具有统计学意义(p>0.05)(表 2).
基于以上实验结果,后续实验均采用体外成熟[0~22) h添加2 nmol/L毛壳素处理方法.
-
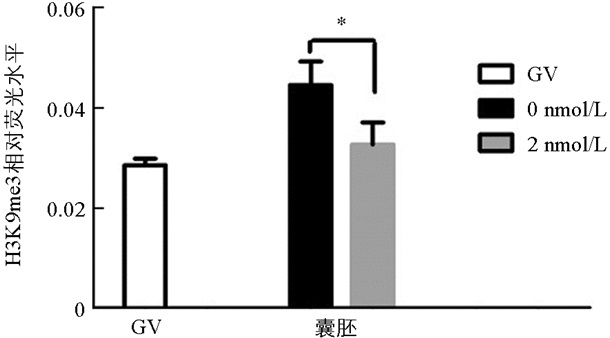
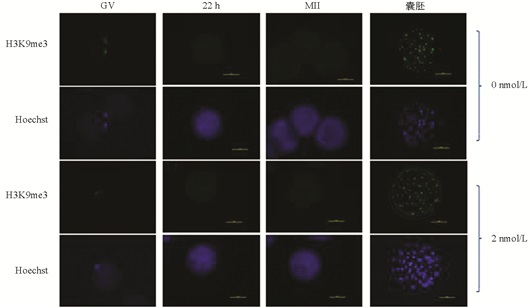
与未处理组相比,H3K9me3在GV期2 nmol/L处理组有明显的表达,差异不具有统计学意义(p>0.05). 随着卵母细胞的成熟进程,H3K9me3表达逐渐减弱,在MI和MⅡ期卵母细胞中未检测到H3K9me3表达. 与未处理组相比,2 nmol/L处理组的囊胚中H3K9me3表达显著降低(p<0.05)(图 1、图 2).
-
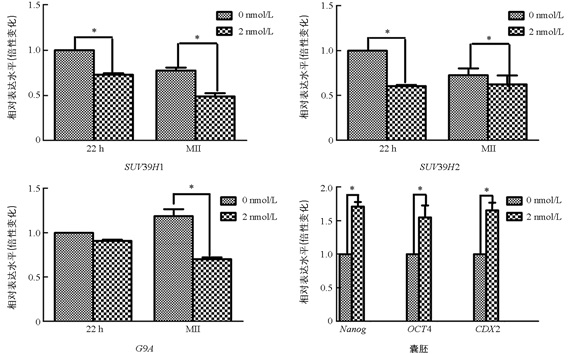
与未处理组相比,2 nmol/L处理组的MI期卵母细胞中的SUV39H1和SUV39H2表达显著降低(p<0.05),两组之间G9A的表达差异不具有统计学意义(p>0.05). 与未处理组相比,2 nmol/L处理组的MⅡ期卵母细胞中SUV39H1,SUV39H2,G9A表达均显著降低(p<0.05);囊胚中Nanog,Oct4,CDX2的表达显著提高(p<0.05)(图 3).
2.1. [0~22) h毛壳素处理对猪卵母细胞体外成熟和早期胚胎发育的影响
2.2. [22~44] h毛壳素处理对猪卵母细胞体外成熟和早期胚胎发育的影响
2.3. 毛壳素处理对猪卵母细胞和早期胚胎中H3K9me3表达的影响
2.4. 毛壳素处理对猪卵母细胞和早期胚胎中重要基因表达的影响
-
近年来的研究表明,胚胎中高表达的H3K9me3是阻碍胚胎发育的关键因素,特别是在合子基因激活(ZGA)过程中[18]. 在ZGA期间,卵母细胞中H3K9me3的异常变化会阻碍转录激活[19],且在核移植胚胎重编程抗性区域(RRR)中检测到高水平的H3K9me3,而体外受精胚胎的RRR基因表达正常. 这些RRRs区域的特征是富含SUV39H1/2并且缺乏DNasr1,这两点都是异染色质的一般特征[20]. 由于H3K9的甲基化与基因的表达抑制密切相关,去除这些表观遗传标记有利于基因的重新激活和胚胎发育. 以往的研究也表明,降低人和小鼠胚胎中H3K9me3水平能提高胚胎的发育能力[10]. 因此,适当降低H3K9me3的表达有利于卵母细胞体外成熟和早期胚胎的发育.
在本研究中,免疫荧光数据显示GV期卵母细胞中H3K9me3的表达水平较高,但MI期和MⅡ期卵母细胞中未检测到该蛋白的表达,发育到囊胚阶段又表现出较高的表达水平,这意味着MII期卵母细胞中H3K9me3的缺失可能正在为通过其他修饰的细胞表观遗传重新编程做准备[21]. H3K9me3的动态表达说明了其在卵母细胞体外成熟和早期胚胎发育过程中具有重要的调控作用.
本实验通过在猪卵母细胞体外成熟过程中添加毛壳素,观察其是否对猪卵母细胞体外成熟和早期胚胎发育产生影响. 结果发现,与未处理组相比,2 nmol/L处理组显著提高了卵母细胞的第一极体排出率、4-细胞率和囊胚率. 在本研究中2 nmol/L处理组囊胚率增加12.92%,表明抑制剂只部分纠正了异常的表观遗传修饰. 通过qRT-PCR和免疫荧光检测发现,毛壳素处理之后显著降低了卵母细胞中SUV39H1/2和G9A的表达,以及囊胚中H3K9me3的表达. 这与Zhang等[22]在羊核移植胚胎中的研究一致. 2 nmol/L毛壳素处理后显著提高囊胚中多能性基因Nanog,Oct4和CDX2的表达,该结果与李海艳[23]对猪孤雌胚胎的研究结果一致. 本实验结果证明适量毛壳素处理有利于猪卵母细胞体外成熟和早期胚胎的发育.
猪卵母细胞中H3K9me3的去除对于正常的ZGA和表观遗传重编程至关重要[18],用毛壳素作为组蛋白甲基转移酶SUV39H1/2和G9A特异性抑制剂,能有效降低H3K9me3的表达水平,最终提高猪体外成熟和胚胎发育效率.



 下载:
下载: