-
开放科学(资源服务)标志码(OSID):

-
粒型是水稻的重要农艺性状,既是产量的重要组成部分,也是外观品质的决定因素之一. 其形态是一个复合性状,主要是由粒长、粒宽、长宽比及粒厚决定,受多基因调控[1]. Gramene数据库(https://archive.gramene.org/qtl/)中收录的水稻籽粒相关性状的QTLs已超过560个,其中部分基因的遗传机理已经被解析. 水稻籽粒大小受复杂遗传网络的调控,涉及G蛋白信号通路、丝裂原活化蛋白激酶(MAPK)信号通路、泛素-蛋白酶体通路、植物激素信号通路、转录调控因子等[2-3].
GS3是通过G蛋白信号通路调控水稻籽粒的QTL,也是在水稻中发现的第1个调控粒长和粒质量的QTL[4],由5个外显子组成,编码产物为跨膜蛋白. 序列分析表明,与小粒品种相比,所有大粒品种的第2外显子均发生无义突变,导致GS3蛋白C端178个氨基酸缺失,造成蛋白翻译提前终止,表明GS3负调控水稻的粒长和粒质量[4-5].
丝裂原活化蛋白激酶(MAPK)是一系列细胞内级联反应的组成部分,可对多种细胞外刺激做出反应[6]. 研究结果显示,MAPK信号通路参与植物生长发育的许多方面,MAPK磷酸酶(MAPK phosphatase,MKP)特异性地将激活的MAPK中的磷酸基去除,从而使其失活[6]. 通过对OsMKK10功能缺失突变体、过表达株系和OsMKK4功能获得突变体的研究表明,OsMKKK10-OsMKK4-OsMPK6级联信号通路调控水稻籽粒大小[7].
GW2编码一个环形E3泛素连接酶,通过泛素-蛋白酶体通路负向调控细胞分裂,影响水稻粒宽和粒质量[8]. OsARF19由生长素和BR共同诱导,通过植物激素调控水稻籽粒大小,OsARF19与BR受体基因OsBR11的启动子结合,直接影响OsBR11的表达,而过表达OsARF19会导致植物表现出矮秆、窄叶、瘪粒和叶片角度增大[9]. GLW7编码植物转录因子OsSPL13,正向调控颖壳细胞的大小,从而造成籽粒的长度、厚度和质量增加[10]. GW8是含有SBP结构域的转录因子,调节水稻籽粒宽度,可直接与GW7启动子结合,抑制其表达[11]. GS2编码OsGRF4,是一种转录调控因子,通过促进细胞分裂和细胞扩张来调控颗粒大小[12].
染色体片段代换系(Chromosomal Segment Substitution Line,CSSL)是研究数量性状的良好材料[13],能将复杂的数量性状分解成含有少量供体亲本代换片段而遗传背景又与受体亲本基本一致的简单性状,从而简化技术难度,提高实验精度. 在前期研究中,我们以日本晴(Nipponbare)为受体亲本,优良恢复系R225为供体亲本,结合多代回交、自交以及分子标记辅助选择(MAS)等方法,培育出了水稻长宽粒染色体片段代换系Z8. 本研究利用日本晴/Z8构建的次级F2分离群体,进行重要农艺性状的QTL定位;同时基于定位结果,结合MAS选育携带代换片段更少、遗传背景更为纯合的次级代换系. 相关研究结果将为水稻粒长、粒宽等重要农艺性状的遗传机理解析奠定理论基础,并提供必要的材料支撑.
全文HTML
-
水稻染色体片段代换系Z8,是以日本晴(Nipponbare)作为受体亲本、优良恢复系R225为供体亲本,经过多代回交、自交并结合MAS法在水稻全基因组水平上选育染色体片段代换系,并运用多态性分子标记从BC2F1开始进行筛选,直到BC3F6选育出来了1个含有14个供体亲本染色体片段的代换系.
QTL定位的群体材料是由日本晴/Z8杂交构建的150株次级F2分离群体.
材料于2019年种植在西南大学歇马水稻研究所基地,用日本晴与Z8进行杂交,收获F1种子后,同年冬季种植于海南基地,收获F2种子. 2020年3月,将日本晴,Z8和F2(150个单株)的种子于西南大学歇马水稻研究基地进行播种;同年4月,以株距和行距为16.67 cm和26.67 cm分别移栽. 2021年3月,将从F2分离群体中选育出来的23个次级代换片段、日本晴和Z8分别种植,每行10株,每个次级代换片段和供、受体亲本各20株. 所有研究材料,包括亲本及相应的分离群体,均参照当地水稻种植与栽培措施进行统一管理.
-
利用分布在水稻全基因组上的429个SSR标记,对日本晴与恢复系R225进行多态性标记筛选,共鉴定出253个多态性SSR. 通过这些多态性SSR结合分子标记辅助选择(MAS),参照王大川等[14]的方法,对BC2F1进行代换片段选择,直到BC3F6选育出含有供体亲本14个代换片段的长宽粒染色体片段代换系Z8. 连续的供体亲本带型标记区间认为是代换片段,参照Paterson等[15]的方法估算代换片段长度,用Mapchart 2.3软件标记代换片段在Z8染色体上的分布.
-
进入成熟期后,齐地收割10株日本晴,10株Z8以及150个F2单株,参照韩龙植等[16]的方法,对粒长、粒宽、长宽比、株高、有效穗数、穗长、一次枝梗数、二次枝梗数、千粒质量、每穗总粒数、每穗实粒数、结实率、单株产量和着粒密度等14个农艺性状进行表型鉴定. 收集所有性状的表型数据,利用Microsoft-Excel 2019进行统计分析与作图.
-
取灌浆期幼嫩的水稻籽粒,利用日立SU350型扫描电镜在-20 ℃环境下观察日本晴和Z8颖壳外表皮细胞并拍照,每个材料重复10次,测量其细胞长度和宽度并对细胞数目进行统计,计算其平均值、标准差,并通过t测验比较Z8与日本晴之间的差异.
-
通过日本晴/Z8构建的次级F2分离群体开展目标性状的QTL定位. 参照McCouch等[17]的方法提取亲本和150个F2单株的DNA,参考王大川等[14]的方法鉴定所有材料的基因型. 结合目标性状的表型鉴定,通过SAS(SAS Institute Inc. 2009)软件,利用限制性最大似然(REML)法进行QTL定位,以p<0.01为阈值判断染色体片段上是否存在QTL位点[18].
-
基于QTL定位结果,并结合表型与基因型,从F2分离群体中筛选出23个单株自交,2021年将自交后代种成株系,并重新编号,其中双片段代换系2个,三片段代换系4个,四片段代换系3个以及五片段及以上的代换系14个,受体亲本、供体亲本以及各个株系各种植20株,利用分子标记辅助选择(MAS)技术进一步对该株系进行筛选,选育单、双和三片段代换系,为研究目标性状上单基因的加性效应和多基因间的上位性效应提供了研究基础.
1.1. 研究材料
1.2. 研究方法
1.2.1. CSSL-Z8代换片段的鉴定
1.2.2. 农艺性状考察与表型鉴定
1.2.3. 籽粒细胞形态学分析
1.2.4. QTL定位
1.2.5. 次级代换片段选育
-
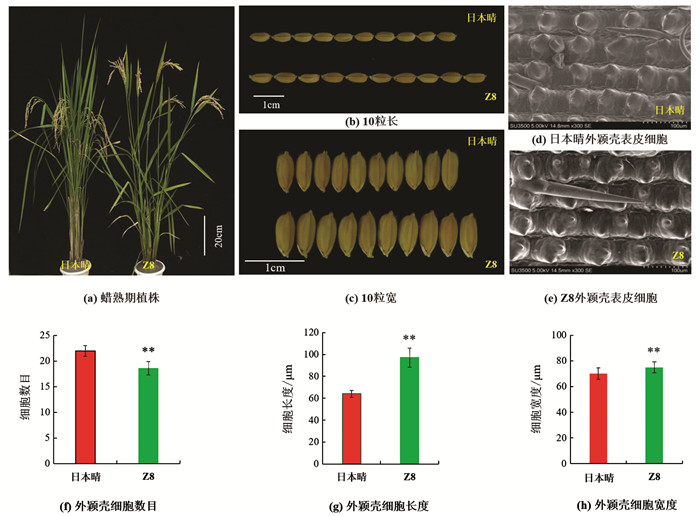
表型鉴定结果显示(表 1),与日本晴相比,Z8的粒长、粒宽、长宽比、株高、一次枝梗数、二次枝梗数、千粒质量、着粒密度和每穗总粒数等9个农艺性状,分别增加了1.18 mm,0.17mm,0.23,8.78 cm,2.92,8.13,3.09 g,32.48和36.95,均显著或极显著高于受体日本晴(图 1a-c). Z8的有效穗数、穗长和结实率比日本晴分别减少了2.80,3.41 cm和21.32%(p<0.01),差异有统计学意义;而每穗实粒数和单株产量差异无统计学意义.
由于Z8的籽粒长度和宽度较日本晴都有所增加,为了解其粒型变化的原因,利用日立SU350型扫描电镜在-20 ℃环境下观察日本晴和Z8孕穗期外颖壳表皮细胞. 结果表明,Z8外颖壳表皮细胞长度、宽度较日本晴分别增加了33.14 μm和5.20 μm(p<0.01),而细胞数目则减少了3.4个(p<0.01),差异有统计学意义(图 1d-h),表明Z8籽粒长度和宽度的增加是由于籽粒外颖壳细胞横向和纵向膨胀导致的.
-
分子鉴定结果显示,Z8共携带了14个来自供体亲本R225的片段,分别位于第1~3,5~9,11,12等10条染色体上,其中第2,7~9等4条染色体上携带2个代换片段(图 2). 估算结果表明,14个代换片段总长度为117.68 Mb,其中最短代换片段长度为1.55 Mb,最长代换片段长度为23.35 Mb,平均长度为8.41 Mb(图 2).
-
利用携带14个供体亲本代换片段的代换系Z8与受体亲本日本晴构建的次级F2分离群体,其农艺性状除株高、有效穗数、单株产量、每穗实粒数和千粒质量的均值较亲本有所增加或降低外,其余均介于亲本之间(表 1,表 2).
由于峰度和偏度是衡量正态分布的两个重要参数,当符合正态分布时,峰度值和偏度值分别为3和0,因此利用日本晴/Z8构建的次级F2分离群体,对其频率分布进行分析. 结果发现,粒型、株高等14个重要农艺性状的偏度值(显示偏离标准正态分布的情况,正值为正偏态,负值为负偏态)均不为0,峰度值均不为3(表 2),表明这些农艺性状均呈现一定的正偏态或者负偏态分布,受多基因控制且基因之间可能存在互作.
-
利用日本晴/Z8构建的次级F2作为定位群体,对在日本晴与Z8之间差异有统计学意义的12个性状进行QTL定位. 结果显示,在10个性状上一共鉴定到33个QTL,其中粒长、粒宽和长宽比各6个;一次枝梗数、二次枝梗数和结实率各1个;每穗实粒数2个;株高和穗长各3个;千粒质量4个(表 3,图 2). 所有QTL对目标性状的表型贡献率在2.04%~88.67%之间,其中有8个QTL对目标性状的表型贡献率超过10%(表 3).
粒长(GL)检测到6个QTL,其中qGL7-1的表型贡献率为14.40%,能够增加粒长0.08 mm,表现为主效QTL. 另5个则均表现为微效QTL(表型贡献率小于10%),其中qGL2-1和qGL2-2分别增加粒长0.06 mm与0.05 mm;qGL1,qGL9和qGL11分别减短粒长0.04 mm,0.04 mm和0.06 mm,表型贡献率分别是7.83%,4.44%,3.53%,3.46%和8.53%.
粒宽(GW)检测到6个QTL,均表现为微效,其中qGW7-1,qGW8-2,qGW8-3分别增加粒宽0.02 mm,0.02 mm和0.01 mm;qGW2-4,qGW6和qGW9分别减小粒宽0.03 mm,0.03 mm和0.02 mm,表型贡献率分别为4.70%,3.41%,3.43%,8.32%,6.81%和4.80%.
长宽比(RLW)同样检测到6个QTL,其中qRLW6的表型贡献率为10.58%,能够增加长宽比0.03,表现为主效QTL. 另5个则均表现为微效QTL,其中qRLW2-1,qRLW2-2和qRLW2-4能够增加籽粒长宽比0.02;qRLW1和qRLW11则相反,均会导致水稻籽粒长宽比降低0.02. 这5个微效QTL对长宽比的表型贡献率分别为5.81%,4.50%,7.68%,7.87%和3.47%.
-
基于QTL定位结果,结合MAS选育了含目标性状QTL的双片段代换系2个(D1-D2),三片段代换系4个(T1-T4),四片段代换系3个(FS1-FS3),五片段代换系5个(FSL1-FSL5),六片段代换系3个(SS1-SS3),七片段代换系3个(SSL1-SSL3),九片段代换系2个(NS1-NS2)以及十二片段代换系1个(TSL1). 虽然Z8的代换片段较多,没有筛选出单片段代换系,不能进行QTL的上位性效应和加性效应研究,但次级代换片段的筛选进一步纯化了材料的遗传背景,为进一步研究数量性状遗传机理提供了丰富理想的材料.
2.1. Z8的表型鉴定与籽粒颖壳的细胞学分析
2.2. Z8代换片段的鉴定
2.3. 日本晴/Z8构建F2次级分离群体表型与频率分布
2.4. 基于Z8的重要农艺性状QTL定位
2.5. 基于Z8的次级代换片段选育
-
本研究鉴定了一个以日本晴为受体亲本,恢复系R225为供体亲本的长宽粒代换系Z8,其携带14个供体亲本代换片段,平均长度为8.41 Mb. 虽然Z8的单株产量无明显变化,但株高、一次枝梗数、二次枝梗数、粒长、粒宽、长宽比和千粒质量较日本晴分别增加了8.78 cm,2.92,8.13,1.18 mm,0.17 mm,0.23和3.09 g(p<0.01),差异有统计学意义. 因此,长宽粒CSSL-Z8作为研究材料对于水稻育种具有重要意义.
根据QTL定位结果分析,在Z8染色体代换片段上所检测到粒长QTLs中,qGL1可能与GW5L为等位基因,通过抑制GSK2的磷酸化活性来正向调节油菜素类固醇(BR)表达,从而负向调节籽粒的大小[19];qGL2-1可能与编码丝裂原活化蛋白激酶OsMKK4的SMG1[20]为等位基因;qGL2-2,qGL7-1,qGL9和qGL11与前人报道的OsmiR156i[21],miR1432[22],DEP1[23]和OsGRF8[24]等基因分别位于同一区间内. 鉴定出的粒宽QTLs中,qGW2-4可能与转录激活因子AFG1等位,通过调节与细胞伸长和增殖相关的几个基因的表达水平进而负向调节籽粒大小[25];qGW6可能与OsNF-YB9为等位基因,主要在发育的胚乳中表达,该基因功能缺失会导致籽粒长、窄、细[26];qGW7-1与芸苔素内酯信号传导的下游信号分子OsBZR1[27]等位;qGW8-3和qGW9可能与已报道的SLG[28]/WTG1[29]和OsSPL18[30]/TAF2[31]等基因位于同一区间内.
此外,鉴定出的控制长宽比的6个QTL,qRLW1,qRLW2-1,qRLW2-2,qRLW2-4,qRLW6和qRLW11是尚未被报道的基因,但这些QTLs分别与前面qGL1,qGL2-1,qGL2-2,qGW2-4,qGW6和qGL11位于相同区间内,可能这些基因在调控粒长、粒宽的同时还影响了籽粒的长宽比,表明基因可能存在一因多效的作用. 对于新发掘的,尤其是主效的QTLs,可进一步精细定位和克隆,为解析这些重要农艺性状形成的分子机制奠定基础.
染色体片段代换系(CSSL)是研究复杂数量性状QTL的主要材料之一,除了从供体亲本引入的片段外,背景与受体亲本几乎相同,具有消除背景对绘图程序影响的独特特征[32-33]. 在本研究中,基于含有14个代换片段的长宽粒代换系Z8构建的次级F2的QTL定位结果,选育出了23个纯合的次级代换系,进一步将控制不同性状以及相同性状的不同基因进行分解,纯化遗传背景,为进一步研究基因间加性效应和上位性效应提供基础,同时为精细定位找明了方向.
-
本研究鉴定了1个以日本晴为受体、优良恢复系R225为供体的水稻长宽粒染色体片段代换系Z8. Z8携带14个来自R225的染色体片段,平均代换长度8.41 Mb. 通过日本晴/Z8构建的次级F2分离群体,共鉴定出33个控制粒长、粒宽等10个农艺性状的QTL,其中有8个QTL对目标性状的表型贡献率超过10%,表现为主效位点. 在QTL定位基础上,结合MAS选育了含目标QTL的双片段代换系2个(D1-D2),三片段代换系4个(T1-T4),四片段代换系3个(FS1-FS3),五片段代换系5个(FSL1-FSL5),六片段代换系3个(SS1-SS3),七片段代换系3个(SSL1-SSL3),九片段代换系2个(NS1-NS2)以及十二片段代换系1个(TSL1). 目标性状的QTL定位以及次级代换片段系的选育,为进一步研究目标性状的遗传机理提供了良好的理论参考和材料支撑.



 下载:
下载:
