-
开放科学(资源服务)标志码(OSID):

-
棉花作为全球重要的经济农作物,是最多的天然纤维来源,被广泛应用于织造行业. 栽培棉花多为异源四倍体,由两个异源二倍体棉属经A基因组与D基因组杂交后染色体加倍形成[1]. 陆地棉(Gossypium hirsutum)作为最重要的异源四倍体棉种占领了超过全球95%的棉花市场. 产量和纤维品质是棉花研究中最受关注的部分[2],但棉花产量与纤维品质呈负相关,难以同时改良[3],是培育优异棉种的巨大挑战. 棉花产量及纤维品质作为数量性状,受多个基因控制. 探究棉花产量与纤维品质的遗传决定因素,利用分子标记检测来为目的基因定位提供依据,对棉花品质改良具有重要意义. 由于陆地棉种内杂交多态性低,因此构建高密度的陆地棉种内遗传图谱是其分子标记辅助选择育种的关键.
构建高密度遗传图谱需要寻找众多的分子标记. 简单序列重复(Simple sequence repeat,SSR)标记分布多且广,实验重复性好且结果可靠,可以在等位基因间显示多个差异[4-5]. 简化基因组测序(Specific locus amplified fragment sequencing,SLAF-seq)是在高通量测序的背景下发展起来的[6-7]. SLAF-seq的3个显著优势是:深度测序保证了基因分型准确;测序成本低;正式测序前利用预测系统提升标记效率. SLAF-seq近年来已被广泛用于单核苷酸多样性(Single nucleotide polymorphism,SNP)检测[8-11].
数量性状位点(Quantitative trait loci,QTL)是在调控生物体数量性状中起重要作用的基因片段在染色体上的位置,数量性状不仅受多个QTL的影响,也与环境互作相关,部分数量性状存在一个主效基因对该性状的控制起主导作用,农作物的产量性状一般为数量性状. 对棉花而言,产量性状、纤维品质和抗逆性等都是数量性状. CottonQTLdb搜集了大量来自全球棉花纤维品质、产量性状、抗病性、耐盐等各类性状的QTL,在release 2.3版本中囊括了截至2018年1月来自156个刊物的4 892个QTL[12-13];Ijaz等[14]总结了2017-2019年多个研究者定位的、与纤维品质相关的稳定QTL.
本研究运用SLAF-seq SNP技术结合SSR分子标记构建高密度遗传图谱,帮助增加遗传图谱的准确性,缩小了QTL长度区间,使QTL定位更加准确高效,可有效推进后续精细定位群体选择和候选基因筛选,为陆地棉产量和纤维品质的分子育种提供参考.
全文HTML
-
杂交母本为渝棉1号,由西南大学棉花研究室自主培育而成,产量高、纤维品质好. 父本为超早3号,由中国农业科学院棉花研究所提供,具有早熟、无腺体、株型紧凑等特点. 重组近交系(RIL)F2:6群体(包含184系)及两亲本材料分别于2019年夏种植于重庆歇马棉花种植基地,2019年冬种植于海南三亚,2020年夏种植于新疆库尔勒和新疆奎屯.
-
提取2019年夏重庆歇马棉花种植基地的棉花植株嫩叶DNA,提取过程参照Zhang等[15]改良的CTAB(Cetyltrimethylammonium Bromide)法. 将提取的184株棉花全套DNA送至北京百迈客生物科技有限公司进行SLAF-seq测序. 参考棉花基因组来自http://ibi.zju.edu.cn/cotton/,利用北京百迈客生物科技有限公司自主研发的基因组酶切软件对DNA进行酶切预测,选择HaeIII+Hpy166II作为最适内切酶组合,经酶切得到的片段范围长度在414~464 bp之间,预测有254 089个SLAF(Specific-Locus Amplified Fragment)标签,经日本晴酶切显示,Control数据双端比对效率为88.74%,酶切效率为84.23%,构建的SLAF库合格.
-
本研究筛选引物包括CCRI,SWU,JESPR,Gh,NAU,BNL,CIR,CGR,HAU,DOW,DPL[16]. 引物CCRI的设计参照亚洲棉基因组[17],引物SWU的设计参照雷蒙德氏棉基因组[18],其他来源于棉花引物数据库http://www.cottonmarker.org. PCR反应体系:1.5 μL 10x Easy Taq PCR Buffer(Mg2+)、0.2 μL 2 mmol/L dNTP、0.3 μL 25 mmol/L Mg2+、0.2 μL 10 mmol/L前引物(pF)、0.2 μL 10 mmol/L后引物、再用ddH2O定容至10 μL. PCR反应流程:预变性94 ℃ 5 min、35个循环(变性94 ℃ 30 sec、退火53 ℃ 30 sec、延伸72 ℃ 30 sec)、延伸72 ℃ 7 min、保温16 ℃ 30 sec. 采用聚丙烯酰胺凝胶电泳法筛选多态性SSR引物,并利用筛选得到的具多态性引物对群体进行基因型检测.
-
将测序结果进行筛选,并将基因型缺失大于80%的标记去除,利用JoinMap 4[19]软件对经SSR分子标记和简化基因组测序得到的SNP标记进行联合分析,作图映射函数为Kosambi函数[20],去除共分离后的位点用于构建遗传图谱,参照Li等[21]测序得到的陆地棉基因组长度计算各染色体的基因组覆盖率.
利用MapQTL©6.0[22-24]对群体进行QTL定位,导入所有环境位点基因型数据和表型数据,选择区间作图法运算得到每个环境QTL的遗传距离,设置似然比对数LOD≥2.5的位点作为确定的QTL邻近标记,LOD值减1的区间作为置信区间,将多个环境检测到的同一性状、同一位置的QTL记为同一个QTL,QTL的命名方式为“q性状-染色体-序号”. 利用MapChart将得到的QTL定位在遗传图谱上. QTL簇的命名方式为“qClu-染色体-序号”.
-
铃质量(BW)、衣分(LP)、籽指(SI)、衣指(LI)按常规方法测定;纤维上半部平均长度(FL)、纤维整齐度指数(FU)、纤维断裂比强度(FS)、纤维马克隆值(FM)、纤维伸长率(FE)送至中国农业科学院棉花研究所进行纤维检测. 各性状间的相关性系数图由R语言绘制:https//www.R-project.org/.
1.1. 材料
1.2. DNA提取与SLAF-seq测序
1.3. SSR分子标记
1.4. 遗传图谱构建与QTL定位
1.5. 数据统计分析
-
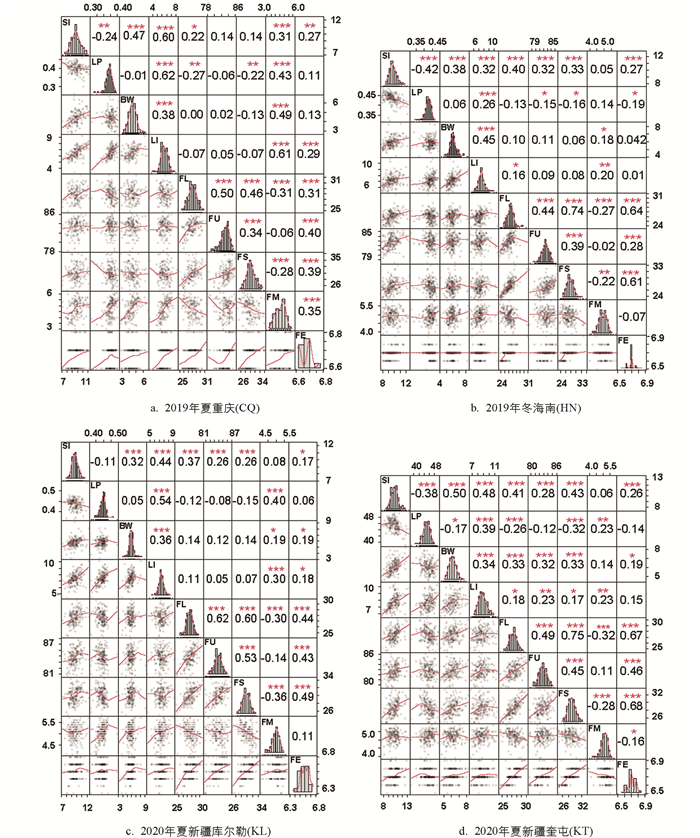
4个环境下9个性状均表现出不同程度的超亲分离现象. 频率分布直方图显示各性状均近似正态分布,且不同环境间性状表现存在差异,结合4个环境的9个表型性状进行方差分析得到F值(表 1),表明这些性状连续分布且易受环境影响,符合数量性状遗传特点. 重组近交系F2:6群体在2019年夏重庆(CQ)、F2:7群体在2019年冬海南(HN)、F2:8群体在2020年夏新疆库尔勒(KL)和新疆奎屯(KT)4个环境中的表型数据如表 2所示. 相关性分析表明,纤维整齐度、纤维断裂比强度、纤维伸长率都与纤维长度呈显著正相关,纤维断裂比强度、纤维伸长率都与纤维整齐度呈显著正相关,纤维伸长率与纤维断裂比强度. 籽指和衣分两两呈显著正相关,纤维长度、纤维断裂比强度都与马克隆值呈显著负相关(图 1).
-
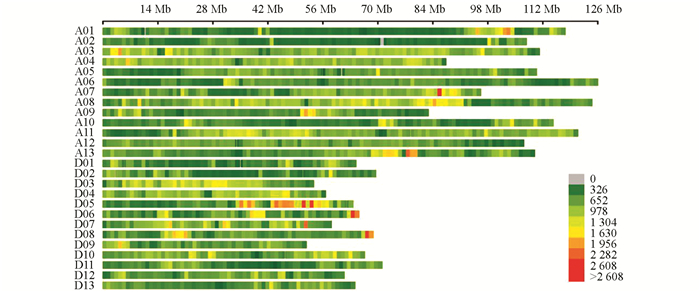
利用3 578对SSR引物对亲本进行多态性筛选得到有多态性的引物145对,多态性比率为4.05%. SLAF-seq测序报告显示共有49 528个SNP标记(图 2). 过滤掉测序缺失率大于80%和颠倒错位的标记. 经SLAF-seq SNP结果和SSR分子标记结果共同分析,最终得到分布于26条染色体的8 020个标记,包括60个SSR分子标记和7 960个SNP标记. 经过分离过滤后,构建了包含2 945个位点,总遗传长度为4 650.71 cM,位点间平均遗传距离为1.58 cM的遗传图谱,物理长度为2 202.95 Mb. 对比TM-1参考基因组的2 240.95 Mb,本文构建的遗传图谱基因组覆盖率为98.30%(表 3).
-
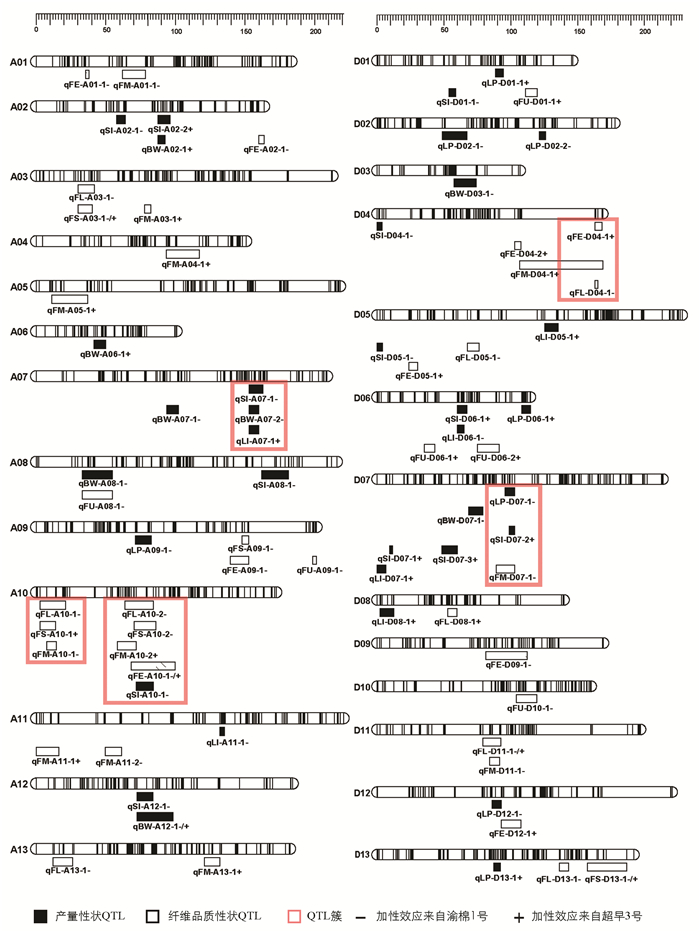
结合高密度遗传图谱与4个环境的产量及纤维品质性状统计数据共同分析,本研究共定位到分布于26条染色体上的76个QTL,包括35个产量性状QTL,41个纤维品质性状QTL,LOD值分布在2.50~7.76之间,解释表型变异值为6.4%~23.4%. 当解释表型变异值大于10%时,视为主效QTL. 加性效应值显示,QTL增效基因来源于渝棉1号的有41个,来源于超早3号的有35个. 有10个QTL在两个及以上环境中被检测到(表 4). 纤维长度性状的QTL qFL-D11-1在4个环境中均被检测到. 在4条染色体上共检测到5个QTL簇,包含17个QTL(图 3).
2.1. 产量和纤维品质表型统计分析
2.2. 遗传图谱构建
2.3. QTL定位
-
分子遗传图谱的构建已经在植物的分子生物学研究中有了广泛的应用[25-27]. 分子标记是构建遗传图谱的前提,在RFLP(Restriction fragment length polymorphism),RAPD(Random Amplified Polymorphic DNA),AFLP(Amplified Fragment Length Polymorphism)等众多的传统标记方式中,本研究选择了广泛应用的SSR分子标记技术,该技术准确度高,可重复利用[28],但工作量大且耗时长. SNP高多态性配合SLAF-seq技术高效且价格相对便宜的优势,成为分子标记的新选择. 在测序结果分析时,筛除了SNP标记中基因型缺失大于80%或者位置明显错误的标记,导致部分区域分子标记存在较大间隙. 本研究结合了SSR分子标记和SLAF-seq SNP技术,且两者相互补充,提高了遗传图谱构建的准确性.
-
高密度遗传图谱是QTL定位的有力保障,多个环境及多个性状的测量使QTL定位更加准确. 在众多QTL定位研究中,定位到的QTL数量差异很大,是因为遗传群体类型不同,亲本遗传背景不同,分子标记类型不同,遗传图谱密度不同,多数表型性状受环境影响大等各类差异所致[29-31]. 研究定位到的QTL增效基因来源不同且QTL加性效应来源也不一致,表明在子代表型统计时出现了超亲分离现象. 本研究结合高密度遗传图谱与多环境性状定位到的76个产量和纤维品质性状QTL,可以为棉花品质精细定位群体的构建提供依据. 在41个纤维品质QTL中,关于纤维长度的QTL qFL-D11-1在4个环境中均被检测到,且其LOD值为所有QTL中的最大值7.76,可为后续棉花纤维品质的图位克隆提供重要参考.
-
QTL成簇是在棉花、水稻、玉米等许多农作物的QTL定位中都存在的现象[32-34]. 关于棉花QTL成簇现象,置信区间内存在“一因多效”现象的解释被广泛认可,Ulloa等[35]指出棉花的重要农艺性状QTL在染色体上表现出高度重组和基因富集的现象;Rong等[36]提出QTL簇的出现还可能是纤维相关QTL代表了一组具有新功能的小基因家族,棉花纤维发育过程的变异涉及复杂的网络或相互作用的基因. 本研究过程中发现当遗传图谱标记间隔较大,定位到的QTL置信区间较大时,QTL之间易出现区间交叉形成QTL簇,因此高密度遗传图谱的构建尤为重要. 本研究在A07染色体上的QTL簇qClu-A07-1包含了籽指、衣分、铃质量3个产量性状的QTL,且加性效应均来自渝棉1号,表明该位点可实现对陆地棉的3个性状同时改良.
3.1. 遗传图谱构建
3.2. QTL定位
3.3. QTL簇
-
本研究构建了一张高密度遗传图谱,包含2 945个上图位点,总遗传长度为4 650.71 cM,图谱物理总长为2 202.95 Mb,覆盖基因组总长的98.30%. 定位到76个QTL,包括产量性状QTL 35个,纤维品质性状QTL 41个,LOD值分布在2.50~7.76之间,解释表型变异率为6.4%~23.4%. 本研究可为后续精细群体构建及候选基因筛选奠定基础,为早熟棉的产量与纤维品质相关育种提供参考.



 下载:
下载: