-
黄花蒿(Artemisia annua)是菊科蒿属1年生草本植物,其药材名为青蒿,已使用近2000年[1].青蒿素(Artemisinin)是从青蒿中提取出的一种倍半萜内酯化合物(Sesquiterpene Lactone),是治疗疟疾的特效药,具有速效和低毒的特点[2-4].青蒿素生物合成起始于植物中普遍存在的法尼烯焦磷酸(Farnesyl diphosphate,FPP),随后在4个特异性青蒿素生物合成酶作用下被转化为青蒿素.编码这4个酶的基因分别是紫槐二烯合成酶基因(ADS)[5]、细胞色素P450单加氧酶基因(CYP71AV1)[6]、青蒿醛Δ11(13)还原酶基因(DBR2)[7]和醛脱氢酶1基因(ALDH1)[8].青蒿素生物合成基因表达水平的高低决定了青蒿素含量的高低,而这些基因表达受到转录因子的严格调控.迄今为止,已经报道了多个调控青蒿素生物合成的转录因子,如AaERF1[9],AaORA[10],AabZIP1[11],AaMYC2[12],AaGSW1[13],AabHLH112[14]和AaPIF3[15]等不同家族转录因子.有些转录因子如AaERF1,AaGSW1,AaMYC2和AabZIP1通过与青蒿素生物合成基因启动子中的相关顺式作用元件(Cis-element)直接结合并激活其表达,从而实现对青蒿素生物合成基因的直接正调控;有的转录因子如AaORA和AabHLH12是通过直接结合转录因子启动子并激活其表达,从而实现对青蒿素生物合成基因的间接正调控.
转录调控是植物次生代谢相关基因表达的主要调控方式之一,转录因子在其中作用明显[16]. bHLH是植物中最大的转录因子家族之一,在发育和生理过程中发挥重要作用,如腺体的形成[17]、激素与光信号转导[18]以及对次生代谢的调节等[19].迄今为止,已发现了4个正调控青蒿素生物合成的bHLH类转录因子,包括AabHLH1[20],AaMYC2[12],AabHLH112[14]和AaPIF3[15],其中AabHLH1,AaMYC2和AabHLH112调控青蒿素生物合成的机制研究较为清楚. AabHLH1和AaMYC2能够直接结合青蒿素生物合成基因启动子中的G-box元件并激活这些基因表达,从而实现对青蒿素生物合成基因的直接调控[12, 20];AabHLH112则是能够结合AaERF1启动子区域的G-box元件并激活AaERF1表达,而AabHLH112并不能结合青蒿素生物合成基因启动子中的G-box元件;过表达AabHLH112能够大幅度提高青蒿素含量,该研究表明AabHLH112是通过直接激活AaERF1表达而实现对青蒿素生物合成的调控[14],因此,AabHLH112对青蒿素生物合成的转录调控是间接调控. AaPIF3同样属于bHLH转录因子家族,能够激活青蒿素生物合成基因表达,在青蒿中过表达AaPIF3能够大幅度提高ADS,CYP71AV1,DBR2和ALDH1的表达水平,从而促进青蒿素生物合成[15].但关于AaPIF3是否直接调控青蒿素生物合成基因并不清楚,同时AaPIF3是否调控AaERF1也未知.
为解析AaPIF3调控青蒿素生物合成的分子机制,采用酵母单杂交(Yeast One-hybrid,Y1H)技术研究了AaPIF3与青蒿素生物合成基因(ADS,CYP71AV1,DBR2和ALDH1)和AaERF1启动子的相互作用,分析了过表达AaPIF3青蒿中AaERF1的表达量,并采用双荧光素酶(Dual-Luciferase,Dual-Luc)报告系统评价了AaPIF3对AaERF1启动子的调控作用,从而明确了AaPIF3转录调控青蒿素生物合成的机制.
HTML
-
实验所用的烟草为本生烟,本生烟种子由西南大学生命科学学院实验室收获保存,本生烟种植于该实验室温室10cm×10cm小方钵内,室温25 ℃,光照周期为光照/黑暗16 h/8 h.本生烟在水肥充足的情况下,生长2~3周即可使用.所用青蒿材料为本团队已经报道的过表达AaPIF3的转基因青蒿和非转基因青蒿[15].
-
酵母单杂交菌株为EGY48,质粒为pB42AD和p178;Dual-Luciferase载体PHB及pGreenII0800-LUC均为该实验室保存[21].
-
酵母单杂交实验可以验证蛋白质和DNA序列能否结合,本实验构建酵母单杂交载体时,将转录因子AaPIF3通过酶切位点EcoRI和XhoI构建到pB42AD载体上,根据G-box元件截段的启动子序列通过XhoI酶切后构建到p178载体上(表 1),包含启动子片段的p178结构如下:pADS(-1 718~-1 127),pADS(-1 144~-539),pADS(-562~-1);pCYP71AV1(-1 150~-840),pCYP71AV1(-844~-560),pCYP71AV1(-286~-1);pDBR2(-1 651~-1 128),pDBR2(-1 219~-448),pDBR2(-467~-1);pALDH1(-2 004~-1 406),pALDH1(-1 490~-934),pALDH1(-1 015~-437),pALDH1(-538~-1);pAaERF1(-1 384~-922),pAaERF1(-994~-448),pAaERF1(-532~-1).通过测序验证重组载体的正确性,杂交实验方法如Zhang等所述[11],将pB42AD-AaPIF3与p178-pADS/pCY71AV1/pDBR2/pALDH1/pERF1的质粒共转化至酵母菌株EGY48,并于Trp,Ura缺陷型固体培养基上30 ℃倒置培养2~3 d后挑取5个酵母单菌落再于Trp,Ura缺陷型固液体培养基中30 ℃,200 r/min培养24 h,离心(12 000 r/min,15 s)收集酵母细胞并用100 μL无菌水重悬,取10 μL重悬菌液滴到X-gal显色培养基上,空的pB42AD及p178质粒作为阴性对照. 30 ℃避光倒置培养48~72 h后观察显色情况并拍照记录.
-
Dual-Luc报告系统被广泛用于研究调控蛋白对目标基因启动子的调控作用,该实验室也曾采用该方法评价AabZIP1,AaAPK1和AabHLH112对青蒿素生物合成基因或相关转录因子启动子的激活活性[11, 14, 22],因此研究将继续采用Dual-Luc评价AaPIF3对AaERF1启动子的调控作用.将AaERF1 (JQ513909)启动子序列克隆并插入到pGreenII0800-LUC质粒的LUC基因上游,得到重组质粒pAaERF1∷LUC作为报告基因,CaMV 35S启动子控制的转录因子(AaPIF3)形成的重组质粒PHB-AaPIF3作为效应基因,CaMV 35S启动子控制的黄色荧光蛋白(YFP)形成的质粒PHB-YFP作为阴性对照.将质粒分别转至农杆菌菌株GV3101中,并将包含PHB-AaPIF3及pAaERF1∷LUC菌株共转化至烟草叶片中,PHB-YFP及pAaERF1∷LUC菌株共转化至烟草叶片作为阴性对照. 48~72 h后取烟草叶片用Dual-LuciferaseR试剂盒(Promega,WI)进行测定,采集同一生长条件下的5个样本进行统计分析.
1.1. 实验材料
1.2. 质粒与菌株
1.3. 方法
1.3.1. 酵母单杂交实验
1.3.2. 双荧光素酶检测实验
-
许多研究表明bHLH类转录因子,如AaMYC2和AabHLH1,通过识别青蒿素生物合成基因启动子区域的顺式作用元件G-box,并与之结合来调控青蒿素的生物合成[12, 20].虽然我们已报道的Dual-Luc实验和转基因结果表明AaPIF3能够正调控青蒿素生物合成基因的表达[15],但并不知道AaPIF3对青蒿素生物合成基因的调控是直接调控还是间接调控. Y1H(酵母单杂交)是常用的研究转录因子和启动子相互作用的技术之一,本研究采用Y1H系统研究了AaPIF3对pADS,pCYP71AV1,pDBR2和pALDH1的相互作用.
分别以ADS基因启动子的3个重叠片段,即pADS(-562~-1),pADS(-1 144~-539)和pADS(-1 718~-1 127),与AaPIF3进行Y1H实验,结果发现在X-Gal培养基上这些转基因酵母并没有变蓝(图 1a).分别以CYP71AV1基因启动子的4个重叠片段,即pCYP71AV1 (-286~-1),pCYP71AV1 (-565~-270),pCYP71AV1 (-844~-560)和pCYP71AV1(-1 150~-840),与AaPIF3进行Y1H实验,结果发现在X-Gal培养基上这些转基因酵母也没有变蓝(图 1b).分别以DBR2基因启动子的3个重叠片段,即pDBR2 (-538~-1),pDBR2(-1 015~-437)和pDBR2 (-1 490~-934),与AaPIF3进行Y1H实验,结果发现在X-Gal培养基上这些转基因酵母也没有变蓝(图 1c).分别以ALDH1基因启动子的4个重叠片段,即pALDH1(-538~-1),pALDH1 (-1 015~-437),pALDH1 (-1 490~-934)和pALDH1 (-2 004~-1 406),与AaPIF3进行Y1H实验,结果发现在X-Gal培养基上这些转基因酵母也没有变蓝(图 1d). Y1H实验结果表明AaPIF3不能结合青蒿素生物合成基因启动子,对青蒿素生物合成基因的转录调控属于间接调控.
-
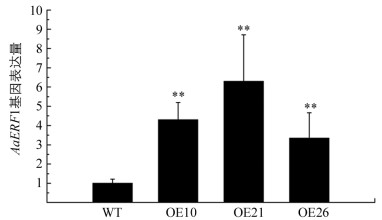
我们先前研究报道AabHLH112能够直接与AaERF1启动子中G-box结合并激活AaERF1表达[14],由于AaPIF3和AabHLH112同属于bHLH转录因子家族,因此推测AaERF1可能是AaPIF3的直接作用靶标基因.通过基因表达量分析发现,过表达AaPIF3的转基因青蒿中AaERF1表达量均比对照显著提高.在转基因株系OE10,OE21和OE26中,AaERF1表达量分别比对照提高了4.30,6.30和3.34倍(图 2).基因表达分析结果表明,AaPIF3对AaERF1的表达具有正调控作用.
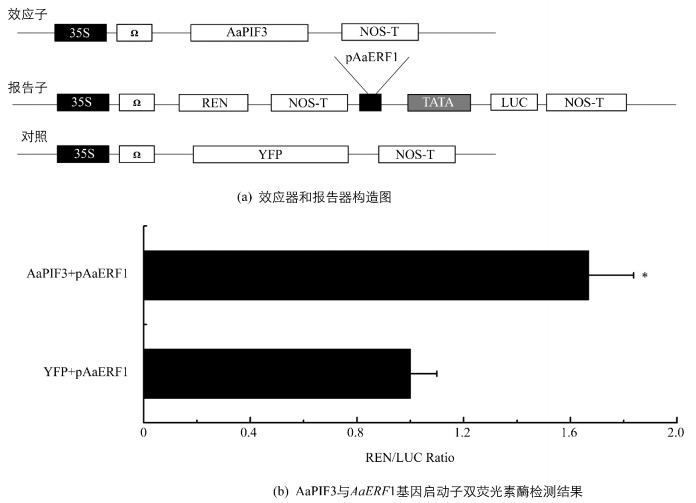
青蒿叶片中AaERF1的表达水平采用Dual-Luc实验进一步研究了AaPIF3对pAaERF1的转录激活作用.在Dual-Luc实验中,构建pAaERF1驱动LUC表达的载体(pAaERF1∷LUC)作为报告子,构建p35S驱动AaERF3表达的载体(p35S∷AaPIF3)作为效应子(图 3a).在共表达pAaERF1∷LUC和p35S∷AaPIF3的烟草叶片细胞中,REN/LUC值为0.060±0.006;在共表达pAaERF1∷LUC和p35S∷YFP的烟草叶片细胞中(对照),REN/LUC值为0.036±0.003(图 3b). Dual-Luc实验结果显示当AaPIF3表达时,pAaERF1活性得到显著增强,为对照的1.67倍(图 3b). Dual-Luc研究结果与AaERF1基因表达分析结果吻合,均表明AaPIF3能够转录激活AaERF1.
-
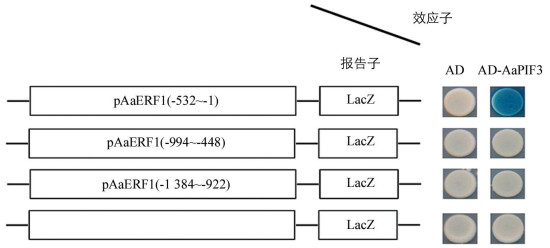
为进一步研究AaPIF3对AaERF1的转录激活作用是否属于直接作用,开展了AaPIF3与AaERF1启动子(pAaERF1)的酵母单杂交研究.将pAaERF1分成3个重叠片段,即pAaERF1(-532~-1),pAaERF1(-994~-448)和pAaERF1(-1 384~-922),与AaPIF3进行Y1H实验,结果发现在X-Gal培养基上只有pAaERF1(-532~-1)截段与AD-AaPIF3的互作片段变蓝(图 4).表明AaPIF3对AaERF1确实有转录激活作用,且通过与AaERF1启动子的G-box作用元件结合调控AaERF1的转录.
2.1. AaPIF3不能结合青蒿素生物合成基因启动子
2.2. AaPIF3能够转录激活AaERF1
2.3. AaPIF3能够结合AaERF1启动子
-
青蒿作为一种富含以青蒿素为代表萜类化合物的药用植物,在不同条件下,需要不同的转录因子来调节包括青蒿素在内的萜类化合物的生物合成[23]. AaPIF3属于含保守HLH结构域的bHLH类家族的转录因子,能与靶基因启动子的G-box顺式作用元件结合[24-25].因此,AaERF1和青蒿素生物合成基因启动子中的G-box顺式作用元件为AaPIF3提供了可能的结合位点[12, 20].酵母单杂交结果表明,AaPIF3直接与AaERF1启动子结合(图 4),但不能与青蒿素生物合成基因的启动子结合(图 1).这些结果表明AaERF1是AaPIF3的直接靶基因,而青蒿素生物合成基因不是.虽然在青蒿素生物合成基因启动子区域也具有G-box,但由于转录因子在结合DNA的时候,核心的顺式作用元件的保守性只是转录因子与DNA结合的前提条件,同时该核心顺式作用元件侧翼序列,甚至处于其远端的其他DNA序列都可能会对转录因子与DNA的结合作用产生影响[26].这也是AaPIF3能够直接调控AaERF1,而不能够直接调控青蒿素生物合成基因的原因.
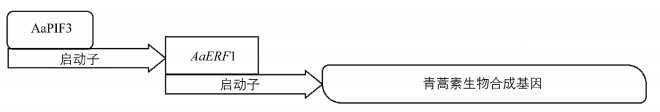
根据以往的研究,转录因子可以直接或间接地调控生物合成基因.转录因子AaERF1,AaMYC2,AabZIP1和AaGSW1通过与青蒿素合成途径关键酶基因的启动子结合来积极调节青蒿素的生物合成[9, 11-12].而另一种ERF类转录因子AaORA间接调控了这些基因,但其过度表达极大地促进了青蒿素在转基因植物中的生物合成[10].双荧光素酶检测为支持AaPIF3在调节青蒿素生物合成中发挥积极作用提供了证据[15].当AaPIF3被表达时,AaERF1的启动子活性显著增加(图 3). AaPIF3的过度表达显著提高了AaERF1的表达水平,而AaERF1通过与ADS和CYP71AV1的启动子结合,直接调控了青蒿素生物合成基因[9].综上所述,我们提出AaPIF3调控青蒿素生物合成的模型:AaPIF3直接结合AaERF1启动子并激活其表达,AaERF1直接激活青蒿素生物合成基因表达,也就是AaPIF3通过直接转录激活AaERF1而间接调控了青蒿素的生物合成(图 5).






 DownLoad:
DownLoad: