-
巨魾Bagarius yarrelli Sykes属于脊椎动物亚门Vertebrate,硬骨鱼纲Teleostei,鲇形目Siluriformes,鮡科Sisoridae,魾属Bagarius.因肌肉呈黄色,俗称“面瓜鱼”、“黄鱼”,在中国仅分布于云南省境内的澜沧江、怒江、元江水系,个体可超过50 kg[1].巨魾与其他鱼类相比,一般捕食小型水生动物[2-3].其肉质风味独特,深受云南版纳、普洱、河口和保山等地人民的喜爱.云南省渔业科学研究院成功地进行了人工繁殖[4].
近年来,鱼类线粒体基因中的12S rRNA基因和16S rRNA基因被用来进行鱼类的遗传多样性分析等[5]对东方鲀属鱼类的Cytb和12S rRNA基因的序列变异情况进行了研究;程国宝等[6]对梭鱼和鲻鱼的16S rRNA和COI基因序列进行了比较分析;唐优良等[7]利用12S rRNA基因对8种笛鲷科鱼类的系统进化进行了分析;童馨等[8]对黄姑鱼的16S rRNA基因序列特征进行了分析,王茜等[9]利用16S rRNA基因研究了尖塘鳢属鱼类的系统进化关系.本文通过对12尾魾属巨魾的12S rRNA,16S rRNA进行测序,序列结果与GenBank里的近缘鱼类进行同源性比较,为阐明巨魾的遗传结构,并进一步开发利用提供理论依据.
HTML
-
用于本实验研究的材料来源于云南省河口县采集的12条巨魾,从每条鱼身上剪取适量的肌肉组织放入EP管中,加入无水乙醇,做好标签后保存于-4 ℃冰箱中.
-
采用杜民等[10]的方法提取巨魾基因组DNA.利用琼脂糖凝胶电泳,在凝胶成像系统下用紫外光照射进行DNA检测,并拍照记录.
-
从GenBank中已经发表的鮡科鱼类的线粒体序列,在保守区采用Primer Premier 5软件设计巨魾特异的12S rRNA和16S rRNA引物共计4对(表 1)[11].
-
巨魾12S rRNA和16S rRNA基因的扩增在PCR扩增仪上进行.其反应体系见表 2.
PCR反应条件为94 ℃预变性4 min,然后95 ℃变性30 s,退火温度依引物而定,72 ℃延伸90 s,共34个循环,最后72 ℃延伸6 min,4 ℃保存.利用1%的琼脂糖凝胶电泳在凝胶成像系统中照相并保存.
-
将检测具有目的片段的PCR扩增产物,用2%的琼脂糖凝胶电泳后在紫外分光光度仪中切下目的片段.用DNA回收试剂盒回收,纯化后克隆到大肠杆菌(具体操作步骤参照回收试剂盒里的说明书).
-
挑取单克隆,在含氨苄青霉素的LB培养基中,37 ℃振荡培养过夜(14~16 h).利用M13通用引物M13(5'-CGCCAGGGTTTTCCCAGTCACGAC-3')和RV-M(5'-AGCGGATAACAATTTCACACAGG-3')进行菌液PCR,筛选出阳性单克隆,送南京金丝瑞生物科技公司测序.
-
处理后得到的12条序列与GenBank里查到的澜沧江巨魾序列利用DNAMAN软件进行对比剪切,得到目的序列.再利用MEGA5.0[12]软件里的Kimura 2-Parameter方法分析12尾巨魾的12S rRNA基因和16S rRNA基因序列碱基含量、遗传距离;利用MEGA5.0软件里的Neighbor-Joining(NJ)法建立9属12种鮡科鱼类的分子系统进化树.
1.1. 实验材料
1.2. 实验方法
1.3. 巨魾12S rRNA基因和16S rRNA基因引物设计
1.4. 巨魾12S rRNA基因和16S rRNA基因的PCR扩增及检测
1.5. 目的片段回收、连接、转化
1.6. 单克隆检测及测序
1.7. 数据处理
-
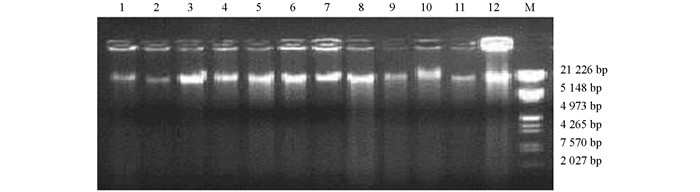
巨魾基因组DNA的提取采用传统的酚-氯仿方法,λDNA /Hind Ⅲ作为相对分子质量标准,提取的DNA片段在21 226 bp左右(图 1).
-
本研究中4对引物的碱基含量不同,退火温度不同.以Baya1F和Baya1R引物对为例,其梯度PCR的退火温度设为12个,依次为53 ℃,53.5 ℃,54.2 ℃,55.1 ℃,56.5 ℃,57.7 ℃,58.6 ℃,59.9 ℃,61.1 ℃,62.1 ℃,62.7 ℃,63 ℃,电泳检测结果见图 2.
-
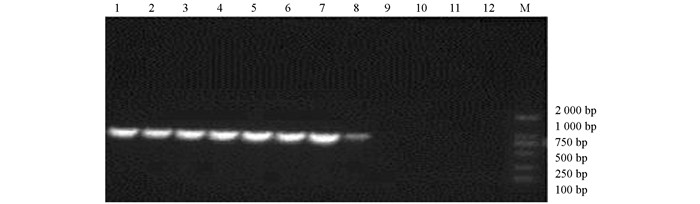
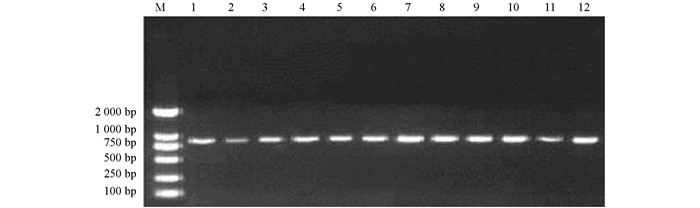
5对巨魾特异性引物对12条巨魾的基因组DNA进行PCR扩增,Baya1F和Baya1R引物对扩增结果见图 3,12个体扩增的目的片段位于750 bp左右(图 3).
-
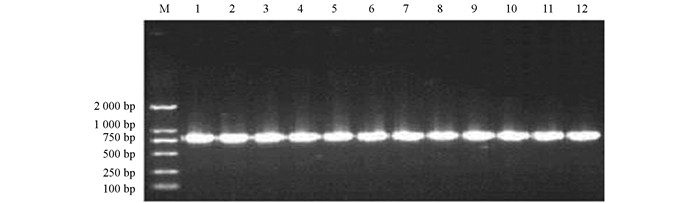
将巨魾12个体PCR扩增结果得到的目的片段利用胶回收试剂盒进行回收,然后连接到载体,转化到感受态细胞进行扩大培养,进行菌液PCR来筛选阳性克隆,Baya1F和Baya1R引物对扩增目的片段胶回收结果见图 4.
-
扩增得到12条巨魾的12S rRNA基因片段经序列比对,长度为954 bp.得到6种单倍型,其中2号、3号、8号、9号个体具有相同的单倍型;样品6号、7号、10号、12号具有相同的单倍型.在6种12S rRNA的单倍型中,简约变异信息位点1个,约占全部位点的0.19%;变异位点有8个,约占全部位点的0.8%;保守位点946个,约占全部位点的99.1%;碱基T,C,A,G的平均含量分别为20.9%,25.7%,32.2%和21.2%,A+T含量为53.1%,高于G+C含量为46.9%(表 3).扩增得到12条巨魾的16S rRNA基因片段长度均为535 bp,得到4个单倍型,其中1号、4号、5号、6号、7号、9号、10号、11号、12号具有相同的单倍型.其中,简约变异信息位点0个;变异位点10个,约占全部位点的0.56%;保守位点532个,约占全部位点的99.44%.碱基T,C,A,G的平均含量分别为20.6%,22.6%,38.3%和18.5%,A+T含量为58.9%,高于G+C含量为41.1%(表 4).
-
利用MEGA 5.0软件中的Kimura 2-Parameter[12]基于12S rRNA基因+16S rRNA基因序列计算12尾巨魾之间的遗传距离,获得12尾巨魾的平均遗传距离为0.002(表 5);转换/颠换的平均值为3.065.
-
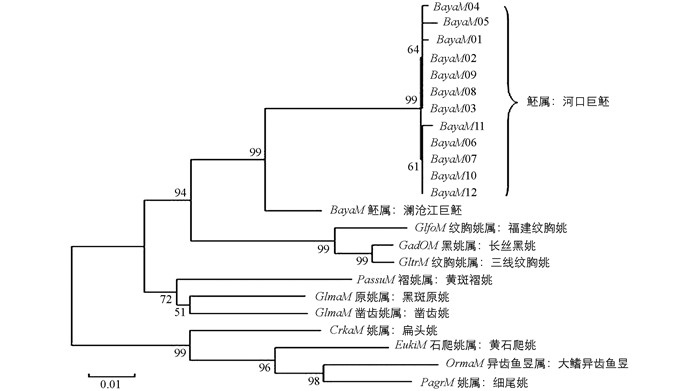
本文利用邻接法(Neighbor-joining,NJ)基于12S rRNA和16S rRNA基因全序列对12种鮡科鱼类构建分子进化系统树(图 5).
由NJ树可知,12种鮡科鱼类分成两大支:即鮡属、石爬鮡属、异齿鱼妟属汇聚成一支,支持率为99%;魾属、黑鮡属、原鮡属、纹胸鮡属、褶鮡属、凿齿鮡属汇聚成一支.黑鮡属与纹胸鮡属是姐妹群,支持率达到99%,其中巨魾单独成一支,支持率达到99%.黑鮡属与纹胸鮡属汇聚成一支.
12种鱼类的科名,种属名见表 6,其中河口巨魾与澜沧江巨魾属于魾属,细尾鮡和扁头鮡属于鮡属,黑斑原鮡和原鮡属于原鮡属.
2.1. DNA检测图片
2.2. 引物退火温度优化结果
2.3. 巨魾PCR扩增结果
2.4. 目地片段回收、克隆
2.5. 多样性分析结果
2.5.1. 巨魾12S rRNA与16S rRNA基因序列碱基组成及单倍型
2.5.2. 12尾巨魾的遗传距离比较
2.5.3. 分子系统进化树
-
实验中扩增得到12条巨魾的12S rRNA基因片段长度为954 bp.序列比对得到同源序列954个位点.其中简约变异信息位点1个,约占全部位点的0.1%;变异位点8个,约占全部位点的0.8%;保守位点946个,约占全部位点的99.1%.扩增得到12条巨魾的16S rRNA基因片段长度为535 bp.其中变异位点有3个,约占全部位点的0.56%;保守位点532个,约占全部位点的99.44%;简约变异信息位点0个.由此可知,两者具有较高的保守性.
12条巨魾的12S rRNA基因序列中T,C,A,G的平均含量分别为20.9%,25.6%,32.2%和21.2%;A+T含量为53.1%,G+C含量为46.9%;12条巨魾的16S rRNA基因序列中T,C,A,G的平均含量分别为20.6%,22.6%,38.3%和18.5%,A+T含量为58.9%,G+C含量为41.1%.以上数据中我们不难看出,12S rRNA基因和16S rRNA基因序列中都是A+T含量略高于G+C含量,本研究的结果与程国宝等[6]在梭鱼和鲻鱼中的研究结果相一致.
12条巨魾的12S rRNA序列及16S rRNA序列中分别出现了6种及4种单倍型,童馨等[8]在100尾黄姑鱼的16S rRNA序列中出现2种单倍型,程国宝等[6]在梭鱼及鲻鱼的16S rRNA序列中分别出现了1种及2种单倍型,表明本研究巨魾的遗传多样性稍微高于黄姑鱼,梭鱼及鲻鱼群体.
-
进行分子系统进化研究中,邻接法(NJ)是最常用的方法[13].由NJ树可知,本研究中所有鮡科不同的种类分成两大分支:即鮡属、石爬鮡属、异齿鱼妟属汇聚成一支,支持率为99%;魾属、黑鮡属、原鮡属、纹胸鮡属、褶鮡属、凿齿鮡属汇聚成一支.黑鮡属与纹胸鮡属是姐妹群,支持率达到99%,其中巨魾单独成一支,支持率达到99%,说明魾属与其他属之间的亲缘关系可能较远.黑鮡属与纹胸鮡属汇聚成一支,它们亲缘关系可能较近.亲缘关系较近的还可能是原鮡属与凿齿鮡属,异齿鱼妟属与鮡属,这与郭宪光[14]等人的研究结果相一致.





 DownLoad:
DownLoad: