-
微RNA(MicroRNA,miRNA)let-7自从被发现后,其在疾病中的作用引起广泛关注.在人类基因组中存在12个由let-7同源基因家族编码的miRNA分子,分别为let-7a-1,a-2,a-3,7b,7c,7d,7e,f7-1,f7-2,7g,7i,Mir-98,其序列具有高度的保守性,在肿瘤、内分泌等疾病中发挥作用.新近的研究报道,let-7家族与肾脏疾病关系密切,let-7a和let-7c在肾透明细胞癌患者中呈低表达,let-7g和let-7i在肾透明细胞癌患者中呈高表达,并与患者的愈后相关联[1-2],Let-7b和let-7c在肾细胞癌患者中呈低表达,并与5-氟尿嘧啶治疗的耐药性产生相关性[3],let-7b靶向GALNT2调控IgA肾病[4],Let-7b通过靶向作用于TGFBR1调控TGF-β1诱导的肾纤维化[5].同时本课题组前期研究通过芯片和real-time PCR检测发现在糖尿病肾病患者中循环let-7a呈低表达,参与疾病的发生[6].因此let-7a可能是肾脏疾病发生发展的因素之一,但其确切机制目前仍不清楚.
miRNA种子序列与靶基因3'UTR序列的互补结合是miRNA发挥生物学功能的重要方式之一,参与疾病的病理生理过程.因此,我们以人肾上皮细胞293T为研究对象,综合运用生物信息学方法和实验室手段,分析let-7a的一个靶基因UHRF1.
HTML
-
Escherichia coli DH5α和人肾上皮细胞293T由本实验室保存.荧光素酶报告载体pmiR-RB-REPORT vector购自广州锐博生物科技有限公司;let-7a mimics,inhibitor及对照购自上海吉玛制药技术有限公司;双荧光素酶报告系统检测试剂盒购自Promega公司;各种限制性内切酶、质粒抽提试剂盒、胶回收试剂盒、逆转录试剂盒购自大连宝生物工程有限公司;Trizol和LipofectamineTM 2000脂质体购自Invitrogen公司;DMEM高糖培养基和胎牛血清购自Hyclone公司;UHRF1抗体和β体actin抗体购自Santa Cruz公司.引物合成和测序由上海生工生物技术有限公司进行.
-
运用microRNA靶基因预测软件TargetScan(http://www.targetscan.org/)、miRTarBase(http://www.mirtarbase.mbc.nctu.edu.tw/)和RepTar(http://bioinformatics.ekmd.huji.ac.il/reptar)预测let-7a的靶基因.在本研究中,至少来源于2个软件的基因才被考虑为let-7a的潜在靶基因.应用Cytoscape[7]的插件BiNGO[8],对预测到的靶基因做GO(Gene Ontology)过度表达分析.从Gene Ontology Consortium[9]的网站下载基因本体和人类基因组GO注释的数据.其中,GO采用最基本的版本(go-basic.obo),过度表达分析采用超几何测试和Bonferroni校正,以全基因组为参考.
-
根据人的UHRF1基因3' UTR序列(GenBank,NM_001048201) 设计上、下游引物:UHRF1-wt-F:5'-CCGCTCGAGTCTCCAAGCACTTCTCGACA-3';UHRF1-wt-R:5'-GAATGCGGCCGCTCTATCTGTGAAAAACATTTATTCTGAGAA-3'.以293T细胞株基因组DNA为模板,按照TaKaRa生物有限公司提供的PCR试剂盒扩增UHRF1基因3'UTR序列.将PCR产物和荧光素酶报告载体pmiR-RB-REPORT vector分别用Xho Ⅰ和Not Ⅰ双酶切,回收目的基因和载体片段,以T4连接酶16 ℃连接过夜12 h并将产物转化感受态DH5α细胞,提取质粒验证获得野生型重组质粒pmiR-RB-UHRF1-3'UTR-wt.根据let-7a种子序列与UHRF1基因3' UTR序列结合位点设计突变引物,UHRF1-3'-UTR-mut上游:5'-GGGTTTGTTAATTTTTCTAATTTTACCAAAGTTTGCAGCCTAGCTTGTCATAAAACAGGGATATTTTAAATCACATACCTGCAGACAAACT-3';下游:5'-AGTTTGTCTGCAGGTATGTGATTTAAAATATCCCTGTTTTATGACAAGCTAGGCTGCAAACTTTGGTAAAATTAGAAAAATTAACAAACCC-3'.测序验证构建质粒.
-
293T细胞接种于100 mL细胞培养瓶,用含10%胎牛血清(Fetal bovine serum,FBS)的DMEM高糖培养基在37 ℃,5% CO2条件下培养,每2~3 d换液. 239T细胞分为下列4个组:let-7a组(细胞转染let-7a mimics)、对照组(细胞转染mimics control)、无处理组及let-7a抑制组(细胞转染let-7a inhibitor).待细胞生长到80%左右,按照脂质体LipofectamineTM 2000试剂盒说明书将let-7a mimics,mimics control和let-7a inhibitor转染至293T细胞,4~6 h后更换新鲜的DMEM培养基,24~48 h后收集各组细胞.
-
将293T细胞按照每孔4×103个细胞种植于96孔板,然后按照Lipofectamine 2000试剂盒说明书将pmiR-RB-UHRF1-3'UTR-wt (200 ng/mL),pmiR-RB-UHRF1-3'UTR-mut (200 ng/mL),pmiR-RB-REPORT vector (200 ng/mL)及let-7a mimics (50 nmol/L)转染至293T细胞.转染48 h后,裂解各组细胞,按Dual Glo Luciferase Assay System说明书操作,以萤火虫荧光素酶活性与海肾荧光素酶活性的比值表示各组细胞的荧光强度.
-
采用Trizol法提取细胞和血液总RNA.采用PrimerScriptTM RT reagent Kit with gDNA Eraser进行逆转录反应. UHRF1引物,上游:5'-ATCTCGGGTGGGGTTTTTCC-3';下游:5'-ATGCTCTAAGGAGTCGGGGT-3'.逆转录反应总体系20 μL,其中5x gDNA Eraser Buffer 2 μL,gDNA Eraser 1 μL,RNA 1 μg,PrimeScript RT Enzyme Mix Ⅰ 1 μL,RT Primer Mix 1 μL,5x PrimeScript Buffer 4 μL,加RNase Free dH2O至20 μL.逆转录反应条件:37 ℃ 15 min;85 ℃ 5 s;4 ℃放置.实时定量反应总体系20 μL,其中SYBR@ Premix Ex TaqII (Tli RNaseH Plus)10 μL,Primer (10 μmol/L) 0.8 μL,RT products 1.6 μL,dH2O 6.8 μL.反应条件:95 ℃ 3 min,95 ℃ 30 s,60 ℃ 30 s,72 ℃ 30 s,共40个循环.每个检验指标做3个复孔.以β-actin作为内参,用2-ΔΔCT法相对定量UHRF1的表达水平.
-
按试剂盒说明书操作提取细胞总蛋白,BCA试剂盒检测蛋白质总含量.根据目的蛋白相对分子质量用10% SDS-PAGE凝胶电泳分离,200V转膜2 h,5%脱脂奶粉室温封闭1 h,按照一抗说明书稀释抗体,4 ℃过夜12 h孵育;TBST漂洗3次,二抗孵育90 min,TBST洗涤3次,ECL化学放光法曝光条带,Quantity One软件采集图像并分析.
-
采用SPSS19.0统计学软件分析,实验数据以±s表示,多组数据使用单因素方差分析,组内比较用Dunnett-t检验.
1.1. 主要材料
1.2. let-7a靶基因生物信息学分析
1.3. UHRF13'UTR野生型与突变型质粒的构建
1.4. 细胞培养和瞬时转染
1.5. 双荧光素酶报告系统检测
1.6. Real-time PCR
1.7. Western blot检测
1.8. 统计学分析
-
运用microRNA靶基因预测软件,得到在3个软件中至少2个软件有交集的let-7a的48个预测靶基因. GO分析结果显示这些靶基因在细胞成分、分子功能、生物过程中参与了诸多生理过程. GO注释有不同的分类,在34条GO注释中生物过程占据30条,表明参与生物过程的let-7a靶基因占绝对优势. KEGG分析结果显示有21条信号通路包含3个或以上的let-7a预测靶基因,这些信号通路主要涉及microRNA在细胞周期、PI3K-AKT、p53等信号通路(图 1).
-
在这48个let-7a预测靶基因中,UHRF1基因的3' UTR含有一个let-7a的结合位点(图 2A),其与let-7a的5'端第2~8位碱基序列,即“种子序列”完全互补,而且该UHRF1基因3' UTR结合位点在多个物种(包括人类、黑猩猩、恒河猴、兔、猪等)的基因组中高度保守(图 2B).提示UHRF1基因3' UTR与let-7a结合位点结合性强,保守度高,可能是let-7a一个较好的靶基因,因此选择UHRF1基因进一步研究.
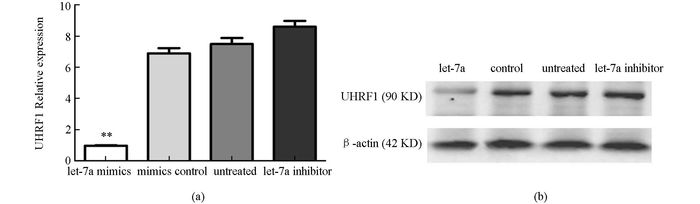
在293T细胞中分别上、下调let-7a的表达,real-time RT PCR和Western blot检测高、低表达let-7a的细胞中靶基因UHRF1的mRNA和蛋白表达.结果显示转染let-7a mimics组UHRF1 mRNA表达水平较对照组及未转染组低,转染let-7a inhibitor组较未转染组高(图 3A),同时转染let-7a mimics组UHRF1蛋白表达水平较对照组及未转染组低,转染let-7a inhibitor组较未转染组高(图 3B),与real-time RT PCR结果相一致.表明let-7a可以负性调节UHRF1基因的表达.
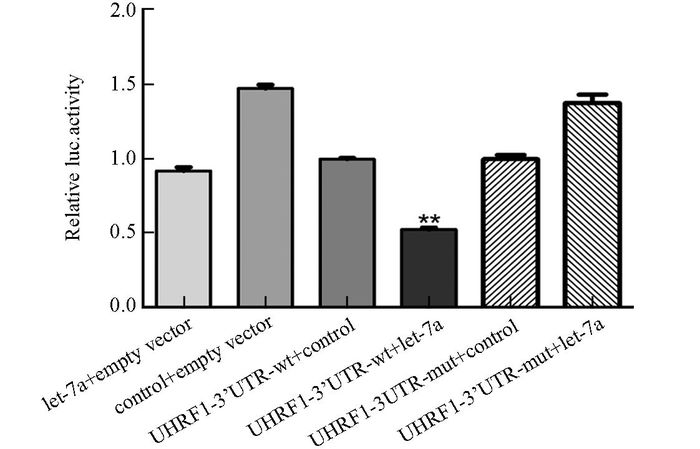
选用包含萤火虫素酶和海肾荧光素酶作为报告基因体系,分别构建了UHRF1-3'UTR-wt和UHRF1-3'UTR-mut的荧光素酶报告载体质粒,测序证实构建成功.荧光素酶活性检测结果显示,过表达let-7a可显著抑制UHRF1荧光酶活性(p<0.01,图 4),提示let-7a可通过与UHRF1 3′UTR结合,负性调控UHRF1的表达,UHRF1是let-7a的靶基因.
2.1. let-7a靶基因生物信息学分析结果
2.2. UHRF1是let-7a的靶基因
-
近年来,随着microRNAs数据的不断增加,这一类小RNA分子为我们提供了一个全新的角度来认识基因和基因表达调控的本质.文献报道miRNA在多种疾病包括肾脏疾病中表达异常,比如Mir-21和Mir-210在肾透明细胞癌表达上调,Mir-141,Mir-200c和Mir-429表达下调[10],MiR-21,miR-192和miR-433在TGF-β1诱导的肾纤维化中具有协同促进作用,而miR-29和miR-200家族则具有抑制效应[11].以上均提示在肾脏疾病的发生发展中,miRNA扮演了重要角色.因此,我们选择进一步研究let-7a在肾病中的生物学功能.
尽管目前有很多microRNA靶基因预测软件,但由于这些软件所采用的计算方式在某些细节上存在差异,导致各个软件的预测结果并不完全一致[12].而且,经生物信息学预测所得的靶基因必须经过实验室研究验证.在本研究中,我们运用microRNA靶基因预测软件TargetScan,miRTarBase和RepTar[13]预测let-7a的靶基因,共有48个至少在2个软件有交集的基因作为let-7a的预测靶基因.进一步运用生物信息学技术对let-7a的预测靶基因进行功能富集性分析. GO分析结果显示这些靶基因在细胞成分、分子功能、生物过程中参与了诸多生理过程. KEGG分析结果显示有21条信号通路包含3个或以上的let-7a预测靶基因,主要涉及microRNA在细胞周期、PI3K-AKT、p53等信号通路.在这些预测靶基因中,UHRF1引起了我们的注意.最近的研究发现,UHRF1通过一种独特的半甲基化CpG结合活性,将DNMT1召集到DNA复制叉周围,维持DNA的甲基化[14],UHRF1通过结合K9甲基化或甲基化DNA,能有效定位到异质染色体上,并在异质染色体的形成与维持中起重要作用[15].
本实验通过生物信息学技术发现UHRF1基因的3'UTR含有一个let-7a的结合位点,其与let-7a的5'端第2~8位碱基序列,即“种子序列”完全互补,而且该UHRF1基因3' UTR结合位点在多个物种(包括人类、黑猩猩、恒河猴、兔、猪等)的基因组中高度保守.因此,UHRF1基因是较好的Let-7a预测靶基因.为证实let-7a与UHRF1是否存在直接靶向作用,我们构建了UHRF1-3'UTR-wt和UHRF1-3'UTR-mut的荧光素报告载体质粒进行荧光素酶活性检测.结果显示,过表达let-7a可抑制UHRF1-3'UTR-wt荧光素酶活性,提示let-7a可与UHRF1 3'UTR上的互补位点结合,从而抑制目的基因的表达.接下来,我们在293T细胞中转染let-7a mimics和inhibitor使之高表达和低表达let-7a,通过Western blot和real-time RT PCR检测结果显示,在293T细胞中let-7a组UHRF1蛋白和mRNA表达量明显降低,而let-7a inhibitor组,UHRF1蛋白和mRNA表达水平上调,表明let-7a可负性调控UHRF1基因的表达,是let-7a的一个靶基因.
本研究从生物信息学手段和实验室证据两方面来探讨let-7a对UHRF1的靶向调控作用,后者建立在前者的基础之上.该研究可为我们提供更丰富的关于let-7a参与转录或转录后调控机制的有关信息,而后续对其靶基因UHRF1的下游效应研究,将为深入认识let-7a的生物学功能提供重要启示.







 DownLoad:
DownLoad: