-
核桃(Juglans regia L.),也名胡桃,羌桃,隶属于胡桃科(Juglandaceae)胡桃属(Juglans),为多年生木本植物.核桃是主要的木本油料树种之一,核桃的叶、花序、果仁、木材以及核桃林本身都具有较高的经济价值,居四大干果之首[1].核桃除含脂肪、蛋白质碳水化合物外,还含钙、磷、铁、硫胺素、核黄素和尼克酸等多种成分,具有很高的营养、药用和经济价值.中国是世界核桃的重要起源地和主产区之一[2],核桃资源非常丰富,包括野生资源、栽培品种资源以及众多地方栽培资源[3].资源是开发新品种的前提,资源分析则是资源研究的基础.资源分析的方法包括形态学、细胞学和分子生物学等,其中分子标记技术的应用越来越多,结果也作为育种的重要参考.对于核桃群体的遗传变异分析已有多年研究,包括形态学指标[4]、同工酶[5]、AFLP[6]、RAPD[7]、RFLPs[8]、SSR[9-10]和ISSR[11-12]等,这些研究覆盖了核桃品种鉴定、亲本鉴定、遗传关系多样性及演化关系等方面.
ISSR(SSR-anchored PCR)是一种新型的分子标记,是由ZIETKEIWITCZ等[13]在1994年发展起来的一种微卫星基础上的分子标记.该技术主要应用于植物资源的分析研究,具有快速、高效、操作简单、扩增产物特异性强、稳定性高和成本低等特点,已广泛应用于梨[14]、樱桃[15]、茶树[16-17]、珙桐[18]、苹果[19-20]和桑葚[21]等树种.核桃种质资源描述在核桃的研究及生产中必不可缺,故而采用一个可靠而高效的分子标记技术应用与资源描述十分重要.从POTTER等对48个核桃品种的ISSR分子标记研究中可见,ISSR技术能很好地区别不同核桃品种,并提供种质资源的起源的证据[12].李国田等[22]利用ISSR技术对新疆核桃元丰、云新核桃、泰山野核桃和陕西核桃4个核桃实生居群共61份种质进行了遗传多样性分析,并探讨了居群间不同种质的遗传变异情况.
重庆秦巴山区由于其特殊的地理环境,植物资源十分丰富,也是核桃的适宜生长区,区域内分布着大量野生核桃群落.该地区栽培核桃的历史悠久,分布也很广泛,如位于大巴山核心区的城口县2011年被国家林业局命名为“中国核桃之乡”,毗邻的巫溪县、巫山县核桃资源也比较丰富,大龄核桃树随处可见,部分资源在产量、抗病性、口感和品质上有较为突出的表现.为了更好地为后续优良品种的选育提供参考,本试验以初步优选的大龄栽培核桃为研究试材,通过ISSR技术分析所搜集的总体核桃资源的遗传多样性,并探讨不同区域核桃实生群体间的关系,了解和掌握大龄栽培核桃的遗传特征,并对后续在育种中的材料利用提供参考依据.
HTML
-
基于调查数据,采集重庆市秦巴山区(包括重庆市城口县、巫山县、巫溪县)经初歩优选的114株核桃树当年生嫩叶片10片以上,置于装有硅胶干燥剂的自封袋中短暂保存于冰盒中,运回实验室后将样品放在-80 ℃超低温冰箱中保存备用.其中用于引物筛选的6份核桃材料分别为ck10,ck26,ck55,ck65,ck66.
-
2014-2015年,于重庆市秦巴山区城口、巫山、巫溪3县核桃集中分布区域,根据产量、品质、果形、成熟期、抗逆性等性状筛选其中一个或数个性状表现较优的30年以上树龄的实生核桃植株.
-
参照ROCHE等[23]改良的CTAB法提取并纯化供试的114份核桃基因组DNA.采用1%的琼脂糖凝胶对所提DNA质量进行检测,并用Thermo NanoDrop 2000将所有样品的DNA质量浓度稀释至约50 ng/μL.
-
ISSR引物参照加拿大哥伦比亚大学(UBC)公布的序列(http://zhidao.baidu.com/question/141178305.html),由上海生工生物工程技术服务有限公司合成. PCR反应在Eppendorf Mastercycler上进行,通过比较和优化,得到最终优化后的体系,具体为:模板DNA 1 μL(50 ng/μL),Taq酶1.0 U,引物0.223 μmol/L,10×buffer 2 μL,MgCl2 1.50 nmol/L,dNTP 0.25 mmol/L,用H2O补至20 μL.反应程序为:94 ℃ 5 min,1个循环;94 ℃ 1 min,50~55 ℃ 30 s(不同引物退火温度不同),72 ℃延伸1 min,35个循环;72 ℃延伸10 min,4 ℃保存.扩增产物用含0.5 μg/mL EB的1%琼脂糖(Invitrogen)凝胶于120 V电压下电泳分离70 min.电泳后在凝胶成像系统上采集图像,记录扩增谱带,保存.
-
电泳结果采取0/1赋值记带,强反应带记“1”,弱反应带重复出现记“1”,弱反应带出现但不重复记“0”,无带记“0”.用NTsys 2.10e软件对材料的遗传相似系数(GS)进行分析,并根据相似系数采用UPGMA进行聚类分析,用POPGENE32软件对供试材料的遗传多样性指数进行分析.
1.1. 试验材料
1.2. 试验方法
1.2.1. 大龄核桃的优选
1.2.2. DNA提取与纯化
1.2.3. ISSR-PCR反应体系优化及引物筛选
1.2.4. 数据统计与分析
-
初步选优结果中城口县共选出74个优良材料,主要集中在厚坪乡、明中乡、高观镇和治平乡等部分乡镇,主要表现为油脂含量高、高产、稳产、抗病以及果大优良性状;巫溪县共选出25个优良材料,集中在双阳乡、乌龙乡以及兰英乡等乡镇,主要优良性状有油脂含量高、高产以及稳产;巫山县共选出15个优良材料,大多选自竹贤乡、曲尺乡及大庙乡,主要优良性状有油脂含量高、高产以及果大.
-
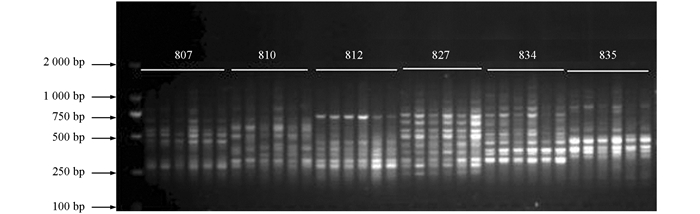
选用6个材料的DNA为模板,从82条ISSR引物中筛选出多态性好,扩增条带清晰的引物用于后续实验.部分引物筛选扩增图谱见图 1.
-
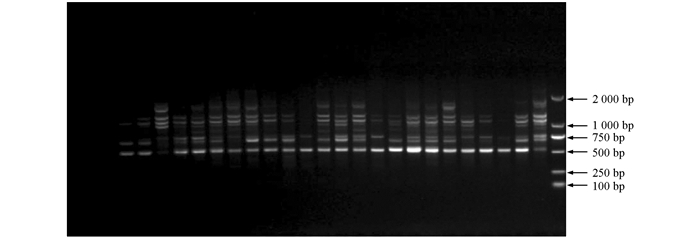
利用筛选出的12条ISSR引物(引物编号分别为807,810,812,827,834,835,840,841,842,844,855,856)对114份供试材料进行基因组检测,12条引物共检测出61个位点,平均每条引物5.08个位点,其中多态性位点58个,平均每条引物扩增出4.83个多态性位点,多态性比率约95.08%,所扩条带分子量分布在2kb~500bp之间(图 2).
利用Popgene.32软件对重庆秦巴山区114份核桃属种质的遗传多样性进行了分析.其平均有效等位基因数(Ne)为1.657 8,Nei'S基因多样性指数(H)为0.373 2,Shannon信息指数(I)为0.547 1.
-
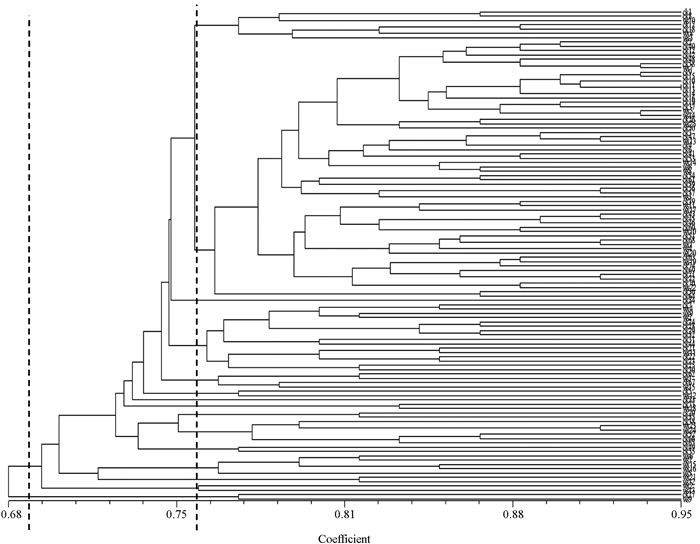
利用NTSYS 2.1软件对114份供试材料的ISSR数据的结果进行聚类分析(图 3).结果显示,供试材料的遗传相似系数的变异范围为0.680~0.936.在遗传相似性系数为0.680处,供试材料共分为两组,第一组材料为ck27和ws9,其余材料归为一组.在遗传相似性系数为0.75处,供试材料分为了13组.
可见,供试的114份秦巴山区核桃种质遗传多样性丰富,不同种质之间的遗传信息有一定的差异.
-
根据采样地点,供试材料可分为城口群体、巫溪群体和巫山群体3个群体.采用Popgene.32软件对3个群分分别进行分析,3个群体的有效等位基因数(Ne)、Shannon信息指数(I)和遗传多样性指数(H)、多态性位点数(NPL),多态性位点比率(PPL)分别见表 1.其中巫溪群体的Ne(1.625 7),I(0.505 6),H(0.347 2),NPL(52),PPL(85.25%)值最低,故巫溪群体间的遗传多样性最低. 3个群体间遗传距离在0.032 7~0.046 2之间,遗传一致度在0.954 9~0.96 7之间.表明群体间相似度高,遗传分化程度小.
UPGMA聚类分析结果显示3个群体被聚为两大类,城口群体与巫溪群体亲缘关系更近,两个群体的遗传距离为0.032 7,遗传一致度高达0.967 8(表 2、图 4). 3个群体间基因流(Nm)为11.197 5,表明不同群体间,基因交流度较高.
2.1. 核桃初步优选结果
2.2. ISSR引物筛选
2.3. 供试材料遗传多样性分析
2.4. 供试材料遗传相似系数及聚类分析
2.5. 不同群体内部及不同群体间的多样性分析
-
我国拥有丰富的核桃育种资源,更好地对这些资源进行鉴定将为核桃育种工作奠定基础.遗传多样性是指种内个体之间或一个群体内不同个体的遗传变异总和,显示了一个物种在特定环境中基因的丰富程度,是植物研究中的重要部分.分子标记方法在多样性研究中的应用较多,ISSR分子标记是一种常用的分子标记,在核桃种质资源的分析中也有报道[22, 24],且与其他标记的研究结果有较高的一致性[1]. ISSR引物在不同物种间通用,且多态性较AFLP,SSR等标记丰富,对遗传背景较为相似的材料更为适合[24].秦巴山区大龄核桃多为栽培类型,其遗传背景可能较为单一.因此,采用ISSR分子标记进行分析是较为合适的,我们的研究结果也证实了这一点.
本研究首次对秦巴山区特殊的生态条件及原始农业管理下的实生核桃种质进行分析.利用ISSR分子标记对3个不同群体的114份重庆秦巴山区大龄栽培核桃进行了分析.结果发现,114份供试材料的遗传相似性系数在0.680~0.936之间.陈良华等[25]对四川干热河谷核桃资源的遗传多样性分析中发现,其遗传相似性系数在0.776 5~0.960 3之间.王红霞等[26]采用AFLP分子标记对普通核桃的遗传多样性进行了分析,结果显示,普通核桃的遗传相似性系数在0.637~0.928之间. CHRISTOPOULAS等[11]对56个世界核桃品种的遗传多样性分析得到,这些品种遗传相似性系数在0.13~0.93之间.可见,重庆秦巴山区农家大龄栽培核桃与其他地区的栽培核桃具有较为相似的遗传多样性,其多样性较为丰富.而不同区域间的相似度较高,这说明不同区域间的基因交流较为频繁,这可能是当地人群自发引种所致.
优良单株的筛选,对后续优良株系的培育有所帮助,其中部分优质、高产单株,如:ck1,ck15,wx1等,可直接用于培育新株系;部分单株,如:CK67,WS11可用作亲本进行品种改良.
由于本研究材料均为初选的在某一方面或多个方面具有优势的大龄核桃单株,这对于了解该区域核桃的起源、分布和资源流向有一定的参考,此外对优质单株遗传指纹图谱的建立有重要的意义,也将对优质单株以及优良品种的进一步选育有所帮助.
-
从现有结果来看,重庆秦巴山区的大龄核桃具有较为丰富的遗传多样性,3个种群之间未产生明显的分化.由于本文所采集材料均为经初选的较优株系,因此,秦巴山区栽培核桃的多样性可能更高.本文利用ISSR技术得到114份核桃资源的指纹图谱,能直接从DNA水平对种质进行鉴定与甄别.本研究可为重庆秦巴山区核桃资源的进一步研究和利用提供参考.






 DownLoad:
DownLoad: