-
禽大肠杆菌病(Avian colibacillosis)是由禽致病性大肠杆菌引起的一种细菌性传染病[1],是危害我国养禽业最重要的细菌性传染病之一[2-3].急性病例引起败血症,亚急性病例表现为心包炎、肝周炎、腹膜炎、气囊炎、肠炎和肉芽肿等症状[1].如果控制不当,会导致很高的发病率和死亡率,给养禽业造成极大的经济损失.对于该病的防控通常采取在饲料或饮水中添加抗菌药物,但抗菌药物的广泛使用使得耐药菌株不断出现,导致该病的防控更加困难.这些耐药菌株随着食物链进入人体,对人的健康造成潜在威胁[4].在人体抵抗力下降,由此类菌株引起发病时容易导致治疗失败.近年来的研究发现,超广谱β-内酰胺酶基因是一类在肠杆菌科细菌中广泛存在的基因群,这类基因都能够产生超广谱β-内酰胺酶(Extended spectrum beta-lactamases,ESBLs),ESBLs能够水解β-内酰胺类抗生素,特别是头孢菌素,是肠杆菌科细菌对β-内酰胺类药物耐药的主要原因[5].本研究分离了某蛋鸡场部分患大肠杆菌病青年蛋鸡的病原,并对病原进行了鉴定,测定了其对多种抗菌药物的敏感性,检测了ESBL基因,为了解该蛋鸡场致病性大肠杆菌的特征积累了数据.
HTML
-
2016年12月,重庆市长寿区重庆长水禽业发展有限公司蛋鸡场出现了初产蛋鸡陆续死亡现象,病鸡表现出精神萎靡、厌食、羽毛松乱、排出绿色或黄白色稀粪等症状.剖检可见肝、脾肿大,肝脏有坏死点,出现腹膜炎、肝周炎、心包炎等一系列病理变化.本实验室对该鸡场送检的8只病鸡进行了病理剖检和病原分离鉴定.
-
革兰氏染液、药敏纸片、肠杆菌科细菌生化编码鉴定管、麦康凯琼脂培养基、MH肉汤培养基购自杭州滨和微生物试剂有限公司,细菌基因组DNA提取试剂盒购自天根生化科技(北京)有限公司,100 bp ladder,2×Taq Master Mix购自北京康为世纪生物科技有限公司. 2日龄健康雏鸡由重庆市垫江县双飞禽业发展有限公司提供.
-
在无菌条件下用灭菌手术刀切开病死鸡的腹腔,剪开心、肝、脾、肺、肾等脏器,用接种环蘸取组织内部后分别划线接种于LB平板、血液平板和麦康凯平板,37 ℃培养18 h后观察菌落形态,革兰氏染色观察细菌形态及染色特性.
-
挑取平板上的单个菌落接种于LB液体培养基,37 ℃ 220 r/min培养6 h.参考Hu等[6]的方法合成大肠杆菌特异的phoA基因片段引物,引物由英潍捷基(上海)贸易有限公司合成.提取基因组后PCR扩增phoA基因片段.反应条件:94 ℃预变性5 min;94 ℃变性15 s,58 ℃退火15 s,72 ℃延伸30 s,25个循环;72 ℃延伸6 min. PCR扩增产物送潍捷基(上海)贸易有限公司双向测序.
-
将2日龄雏鸡随机分为22组,每组2只.每组分别灌服新鲜培养的分离菌菌液2 mL(含活菌8×109 CFU),共21组,对照组灌服2 mL PBS.观察雏鸡的发病和死亡情况,及时剖检死亡鸡,并进行细菌分离.
-
经鉴定的病原菌接种15种肠杆菌科细菌生化鉴定管,分别于37 ℃培养24 h和48 h后观察生化反应.
-
根据美国临床和实验室标准协会的药物敏感性试验方法,以大肠杆菌(ATCC25922)为质控菌株,将合适浓度的各大肠杆菌分离株菌液涂布MH平板,药敏纸片贴于平板表面,37 ℃培养16~18 h,测量抑菌圈直径.
-
PCR扩增分离菌株中的8个ESBL基因blaTEM[7],blaCTX-M[7],blaSHV[8],blaOXA-2[9],blaOXA-10[10],blaVEB[11],blaPER[12]和blaGES[13],预计扩增片段长度分别为931 bp,868 bp,909 bp,914 bp,478 bp,720 bp,927 bp和864 bp.引物由英潍捷基(上海)贸易有限公司合成.
1.1. 病例描述
1.2. 材料与试验动物
1.3. 细菌的分离培养
1.4. 大肠杆菌phoA基因的PCR扩增
1.5. 致病性试验
1.6. 生化试验
1.7. 药物敏感性试验
1.8. ESBL耐药基因的检测
-
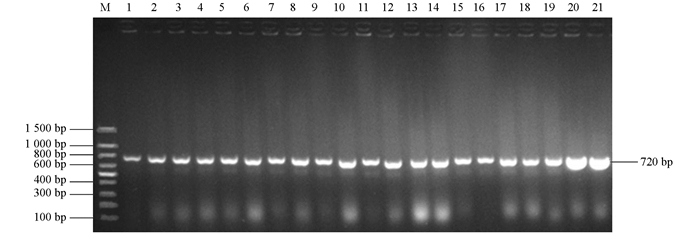
在8只鸡的心血、肝、脾、肺、肾中共分离到21株细菌.经PCR鉴定,所有分离菌均能扩增出720 bp的phoA基因片段(图 1).经测序,PCR产物的核苷酸序列与phoA核苷酸序列相同,证明扩增的片段为phoA基因片段.在麦康凯平板上形成表面光滑湿润、边缘整齐的桃红色菌落,革兰氏染色后镜检可见细菌为短杆状的革兰氏阴性细菌.因此,所分离菌株均为大肠杆菌.
-
分离的21株菌分别接种2日龄雏鸡. 10只鸡24 h内死亡,12只鸡48 h内死亡,15只鸡72 h死亡,5只鸡未死亡.死亡的鸡在6 h内,未死亡的鸡于灌服菌液5 d时进行剖检,所有鸡均出现典型的大肠杆菌病的症状和病变,如肝脾肿大、腹膜炎、心包炎等,从病死鸡的心、肝、脾、肺、肾等实质器官均能分离出与接种菌一致的病原菌.灌服PBS的对照组雏鸡没有任何病变,在实质脏器不能分离到细菌.
-
生化试验鉴定结果表明(表 1),所有分离菌株均能运动(半固体琼脂),能利用赖氨酸、鸟氨酸、尿素、葡萄糖、山梨醇、木糖,一些菌株还能利用尿素、葡萄糖、山梨醇和木糖产气;除了2株菌,其余菌株均能发酵棉子糖;所有菌株均不能利用硫化氢、苯丙氨酸、葡萄糖酸盐、蛋白胨、葡磷胨、枸橼酸盐,只有1株细菌能够利用侧金盏花醇.
-
本试验测定了分离菌株对30种抗菌药物的敏感性.由表 2可知,所有菌株对青霉素G、甲氧嘧啶、复方新诺明、四环素和万古霉素耐药,对阿米卡星敏感;80%~99%的菌株对阿莫西林、卡那霉素和氯霉素耐药,对头孢哌酮、头孢曲松、头孢他啶、妥布霉素敏感;50%~79%的菌株对环丙沙星、恩诺沙星、诺氟沙星、氨苄西林、头孢噻吩、氟苯尼考、红霉素耐药.总体来看,这些临床分离株对测定的喹诺酮类、青霉素类、磺胺类、酰胺醇类药物及四环素和万古霉素耐药较为严重,对测定的头孢菌素类、氨曲南、氨基糖苷类药物及强力霉素和阿奇霉素相对较为敏感.
-
根据美国临床和实验室标准协会大肠杆菌ESBL表型的筛选和确证试验,通过初筛(测定菌株对头孢他啶、头孢噻肟、头孢曲松和氨曲南的敏感情况)可知,所有菌株均有可能产ESBLs,经确证试验鉴定,共有9株菌一定能产ESBLs.
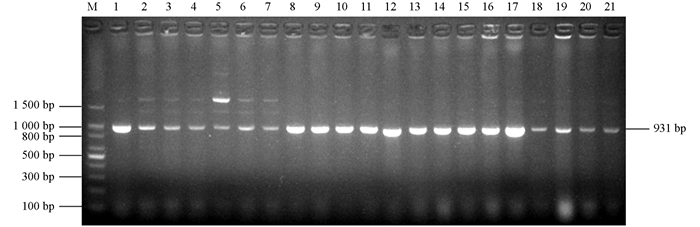
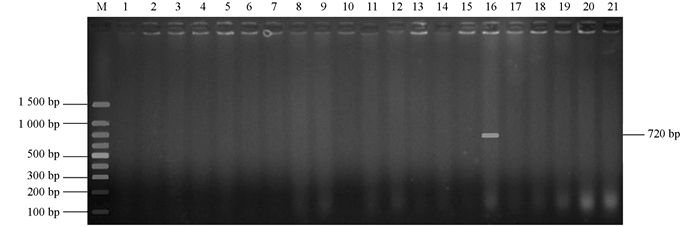
为了在分子水平鉴定出ESBL细菌,扩增了所有8个ESBL大类基因型blaTEM,blaCTX-M,blaSHV,blaOXA-2,blaOXA-10,blaVEB,blaPER和blaGES.所有菌株均携带blaTEM基因(图 2),1个菌株还携带blaOXA-10基因(图 3),该菌株的ESBL基因型为blaTEM+blaOXA-10,没有菌株携带blaCTX-M,blaSHV,blaOXA-2,blaVEB,blaPER和blaGES基因.将图 2中1~7泳道1 500 bp左右的条带切割后测序,表明为非特异性条带.
2.1. 细菌分离情况
2.2. 分离菌致病性试验
2.3. 生化试验结果
2.4. 药敏试验结果
2.5. ESBL表型及基因型鉴定结果
-
本试验通过细菌和菌落形态观察、分子生物学鉴定、生化试验和动物回归试验,确定从病鸡分离的21株病原菌为大肠杆菌.虽然这些病原菌分离自同一个养殖场,但不同的菌株通常具有不同的生化反应特性,说明这些细菌来自不同的克隆株.
抗菌药物被世界许多国家用于细菌性传染病的防制及作为饲料添加剂提高畜禽生长速度.在抗菌药物的选择压力下,耐药菌在养殖场内的比例越来越高,并通过食物链、环境和直接接触传递给人类[14-15],耐药基因也通过动物源性细菌传递给定居于人类的细菌[16].本研究的试验结果也说明耐药菌,特别是多重耐药菌占分离菌株的大部分,细菌耐药非常严重.虽然我国已经有动物禁止或限制使用的药物,但在养殖场中仍然能够分离到禁用药物的耐药菌株,这可能由3种原因造成:①在药物禁止使用之前已经产生了针对该药物的耐药菌株;②可用药物和禁用药物有相同的耐药基因,如floR基因是氯霉素和氟苯尼考共有的耐药基因[17],分离的菌株存在耐氯霉素的floR基因,可能是由于氟苯尼考的使用造成的;③环境中的耐药菌株与养殖场非耐药菌株之间耐药基因的交换.蛋鸡在产蛋期禁止使用抗菌药物,但本试验的分离菌株仍然存在非常严重的多重耐药菌,其原因可能是在该养殖场早期使用过抗菌药物,且该养殖场的菌株和外界耐药菌株耐药基因的交换所致.
近年来鸡源产ESBL大肠杆菌不断被发现并呈逐年上升趋势.细菌对β-内酰胺类抗生素耐药主要是由于产生β-内酰胺酶,使β-内酰胺环的酰胺键断裂而失去抗菌活性.大肠杆菌和肺炎克雷伯菌是最常见的产ESBL细菌.针对ESBL在肠杆菌科广泛存在的情况,本研究鉴定了分离菌株的表型和基因型.结果显示所有的分离菌均存在ESBL,除了一株为TEM+OXA-10外,其他基因型为TEM.这说明产ESBL大肠杆菌是该养殖场的优势致病性大肠杆菌.出现大量产ESBL大肠杆菌的原因可能是ESBL基因非常容易传播,即使在没有选择压力的情况下也可以很容易在细菌间传播[18-19].
本研究通过对该养殖场病鸡的病原分离鉴定和病原的药物敏感性测定,初步摸清了该养殖场致病性大肠杆菌的耐药情况,为养殖场进一步采取综合措施防控该病提供了参考依据.





 DownLoad:
DownLoad: