-
茶树(Camellia sinensis)是世界上三大饮料作物之一,具有重要的经济、保健和生态价值.目前茶树的分布最北界限达北纬42°,最南达南纬33°,遍及四大洲的50多个国家[1].茶树的原产地位于我国云贵高原以大娄山脉为中心的地域,其中云南普洱茶和贵州绿茶均以独特的风味品质而被大家熟知[2-3];茶树原产地得天独厚的自然环境和悠久的种茶历史造就了丰富的古茶树资源,由于其具有广泛的遗传变异和大量原始的优良基因,近年来愈来愈受到茶叶界的关注[4].贵阳花溪古茶树栽培历史悠久,分布广泛,资源丰富,仅久安乡境内就具有保护价值的古茶树54 000多株,占地266.67余hm2,其中2000年以上的古茶树有19株,1 000~1 500年之间的古茶树有1 450株.因此该区域的古茶树资源是开展茶树起源、演化、种质创新和新品种选育等研究的天然宝库.随着分子生物学发展,以DNA分子标记为标志的基因组研究进展迅速,RAPD[5-7],RFLP[8],AFLP[9-10],SSR[11-12],ISSR[13-14]和EST-SSR[15]等标记已用于茶树种质资源和品种鉴别、遗传多样性、亲缘关系和遗传演化等研究.单核苷酸多态性(Single nucleotide polymorphism,SNP)技术具有自动化程度高、通量大、速度快、易于标准化操作、适合大规模研究及基因分型等优点.李志远等[16]搜集生产上主要甘蓝品种,利用重测序数据与参考基因组进行比对开发了2.54×106个SNP标记.马宇等[17]利用SLAF-seq测序技术获得1 105 347个向日葵SLAF标签,其中多态性SLAF标签共有86 985个,获得了414 692个群体SNP标记.李敏等[18]采用SLAF-seq技术获得了277 333个乔木柳SLAF标签,其中多态性标签99 526个,开发了9 488个SNP位点.刘丽华等[19]仅需14个SNP位点就可将378份测试小麦中的99.5%的材料区分开.本试验拟采用SNP技术,对贵阳花溪古茶树进行分子标记开发,获得全基因组范围内的分子标记,了解贵阳花溪古茶树的遗传多样性、群体结构和遗传进化,为茶树特异SNP标记开发、资源鉴定分析、高密度遗传连锁图谱构建和重要农艺性状的关联分析等奠定基础.
HTML
-
供试材料来自贵州省不同地区的48份典型古茶树,其中30份古茶树材料来自贵阳花溪,其余18份古茶树材料来自贵州其它不同地区,古茶树的来源及树型的详细信息见表 1.
-
古茶树叶片基因组的提取采用CTAB法[20],其具体方法为:称取新鲜幼嫩叶片0.2 g,液氮研磨至粉末,移入盛有600 μL 2% CTAB提取液的离心管中,混匀;65 ℃水浴1 h;冷却至室温后,加入600 μL的氯仿:异戊醇(V:V=24:1),上下混匀10 min;4 ℃ 13 000 r/min离心10 min;吸取上清液,放入600 μL异丙醇的离心管中,轻轻混匀;4 ℃ 13 000 r/min离心10 min;弃上清,加入1 mL 70%乙醇清洗DNA沉淀,弃上清,用吸水纸吸干管口水分,气干;加入50 μL已灭菌的超纯水,溶解及混匀DNA.所提取DNA经超微量分光光度计检测质量和浓度后,-20 ℃保存备用.
-
利用SLAF-seq(Specific-locus amplified fragment sequencing)技术对供试材料进行分子标记开发,SNP标记的开发是以每个SLAF标签中深度最高的序列类型作为参考序列,利用bwa[21]将测序reads比对到参考序列上,并使用GATK[22]和samtools[23]两种方法开发SNP,以两种方法得到的SNP标记交集作为最终可靠的SNP标记数集,最终获得全基因组范围的具有代表性的高质量SNP标记.
-
通过MEGA5软件[24]和Neighbor-joining[25]算法进行古茶树的进化分析;通过Admixture软件[26]分析古茶树样品的群体结构;通过Cluster[27]软件进行主成分分析.
1.1. 材料
1.2. 方法
1.2.1. 古茶树基因组DNA提取
1.2.2. 古茶树基因组SNP标记分析
1.2.3. 数据分析
-
本试验利用生物信息学分析古茶树样品重测序数据,共开发1 656 258个古茶树SLAF标签(特异长度的DNA片段),古茶树样品平均测序深度为24.21x,其中多态性SLAF标签有462 897个,共得到2 690 638个群体SNP标记,根据完整度>0.8,MAF>0.05过滤,共获得283 376个高一致性的群体SNP标记(表 2).
-
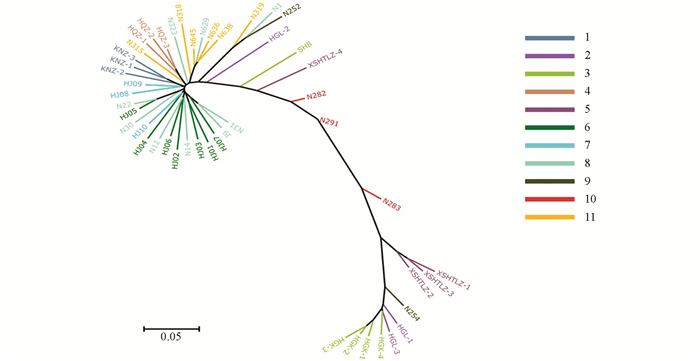
进化树用来表示物种之间的进化关系,根据各类生物间亲缘关系的远近,把各类生物安置在有分枝的树状图上,简明地表示生物的亲缘关系和进化历程.本试验基于SNP标记,对48份贵州古茶树资源进行了进化分析,计算得到古茶树的进化树(图 1).
从进化树可以看出,古茶树资源间存在丰富的遗传变异,来自相近地方的古茶树在进化树上基本处于相近位置,但是贵阳花溪古茶树存在着广泛的遗传差异.位于进化树最下端的古茶树资源分别为HGK-1,HGK-2,HGK-3,HGK-4,HGL-1和HGL-3(来自贵阳花溪高坡乡).顺着进化树向上为来自贵州普定的N254、来自贵州习水的XSHTLZ-1,XSHTLZ-2和XSHTLZ-3、来自贵州务川的N283,以上古茶树资源的树型均为乔木.顺着进化树继续向上的古茶树资源树型开始变为灌木,最上端的古茶树资源分别为N31,J9,HJ07,HJ01,HJ03,N14和HJ02等,均来自贵阳花溪,由此可见,花溪古茶树分布非常广泛,从进化树下部到上部均有分布.
-
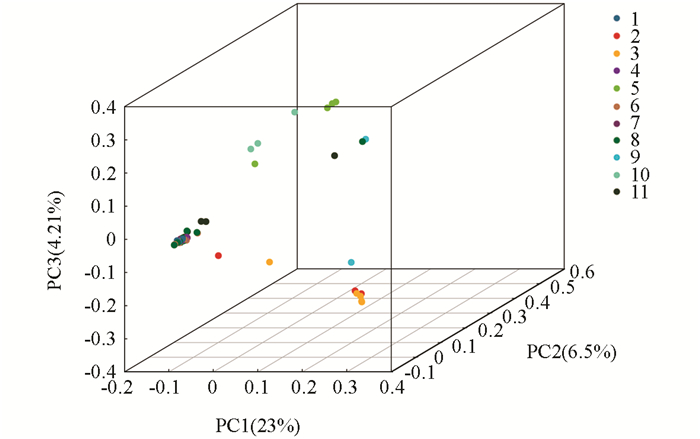
本试验基于SNP标记,通过Cluster软件,对供试古茶树资源进行了主成分分析(Principal component analysis,PCA),得到48份古茶树的主成分聚类图(图 2).从图 2可以看出,样品间存在广泛的差异,来自同一地方的茶树基本上聚在一起,说明它们之间的亲缘关系比较近,但是花溪古茶树的遗传差异最大,分布范围广泛,该结果和前面进化树的分析结果是一致的.
-
群体遗传结构分析能够提供个体的系统来源及其组成信息,是一种重要的遗传关系分析工具.本试验分别假设48份古茶树资源的分群数(K值)为1~10进行聚类,当K为2时,CV值(变异系数)最小,根据CV值来确定分群数为2(图 3),可将48份古茶树资源分为2个类群——灌木型古茶树(类群1)和乔木型古茶树(类群2);类群2中古茶树包括11份材料(HGK-1,HGK-2,HGK-3,HGK-4,HGL-1,HGL-3,XSHTLZ-1,XSHTLZ-2,XSHTLZ-3,N254和N283),属于高大的乔木型古茶树,其余37份古茶树材料皆属于类群1,它们是较低矮的灌木型古茶树,这与进化树的分析结果一致.
2.1. 古茶树SLAF标签与SNP标记开发
2.2. 古茶树的系统进化树分析
2.3. 古茶树主成分分析
2.4. 古茶树群体结构分析
-
茶树世代周期长,自交不亲和,遗传组成高度异质杂合,利用常规育种方法培育新品种费时耗力[28].近年来,随着分子生物学的发展,以DNA分子标记为标志的基因组研究进展迅速,已广泛用于茶树种质资源和品种鉴别、遗传多样性、遗传演化及稳定性等研究[4].金惠淑等[29]利用RAPD技术研究了中国、韩国和日本的46个茶树样品的遗传多态性,19个随机引物扩增出200条带,发现中国茶树品种的遗传多样性高于韩国茶树品种. Lai等[30]利用53个RAPD和56个ISSR标记对台湾野生茶和栽培品种进行了研究,结果表明,台湾本土野生茶与阿萨姆类型亲缘关系更为接近,在所研究的3个居群中,台湾野生茶的遗传多样性最为丰富.牛素贞等[15]利用29个EST-SSR标记对贵州25份地方茶树群体种和3份育成品种的遗传多样性、亲缘关系和遗传分化进行了分析,发现贵州地方茶树品种资源间的遗传差异较大,黔南地区茶树资源遗传多样性程度高于黔北地区,两地区遗传分化较高,基因交流频率较低.本试验开发了1 656 258个古茶树SLAF标签,其中多态性SLAF标签有462 897个,共得到283 376个高一致性的群体SNP标记.从基于SNP标记获得的进化树可以看出,贵州茶树样品间存在丰富的遗传变异,进化树最下端是来自花溪高坡的乔木型古茶树(HGK-1~HGK-4等),顺着进化树向上依次是来自普定、习水、务川、花溪久安、安顺、开阳和花溪的古茶树,由此可得出花溪古茶树分布非常广泛,从进化树下部到上部均有分布.通过PCA分析发现,来自同一地方的古茶树在PCA聚类图上基本处于相近位置,但是花溪古茶树存在着广泛的遗传变异,获得了与进化树一致的结果.
Matsumoto等[8]利用PAL(丙氨酸解氨酶)cDNA作为RFLP探针,对29个日本绿茶栽培品种和地方品种的遗传变异进行了研究,将日本绿茶品种分为5类. Kaundun等[6]利用SSR技术将24份茶树资源分为Assamica和Sinensis两个类群. Mondal[11]利用SSR引物对25份茶树资源进行分析,将所有资源分为禅叶型、阿萨姆型和中国型3个簇. Wachira等[31]采用RAPD和AFLP两种标记,将来源于印度、斯里兰卡、中国、日本、越南和肯尼亚等48个品种划分为阿萨姆变种、中国变种和栽培型与野生型茶树品种3个类型. Balasatavanan等[32]应用AFLP标记将49个茶树品种资源划分为阿萨姆、中国和阿富汗过渡型3个类型.本试验通过古茶树的群体结构分析,清晰地把古茶树资源分为2个类群——乔木型古茶树与灌木型古茶树,其中11份材料属于高大的乔木型古茶树,其余37份材料为较低矮的灌木型古茶树.陈亮[5]认为茶组的系统演化途径由乔木向灌木演化,本试验通过基于283 376个SNP标记的古茶树进化树分析,发现乔木型古茶树分布于进化树下部,而灌木型古茶树位于进化树上部,证实了茶树是由乔木型向灌木型进化的.






 DownLoad:
DownLoad: