-
开放科学(资源服务)标志码(OSID):

-
湿地在生态恢复中发挥着重要作用. 湿地具有多种生态功能,包括元素生物地球化学循环、作为有机碳库减少温室气体如CO2,N2O和CH4释放等. 人工湿地具有较好的景观效果和污染物处理效率,因而在生活污水处理中得到了广泛的应用[1]. 湿地影响着细菌多样性、丰度和均匀度[2],同时,细菌的种类与群落结构对于湿地生态功能至关重要. 目前对湿地细菌群落及其功能的研究主要集中在红树林湿地的硫代谢微生物[3]以及影响红树林湿地细菌区系的因素[4],杭州西溪湿地公园的细菌多样性、组成和类型,用标记基因nrfA研究上海湿地公园细菌硝酸异化性还原为氨(Dissimilatory nitrate reduction to ammonia,DNRA)涉及的高丰度微生物. 有研究[5]发现细菌丰度从高到低依次为湖球菌属(Lacunisphaera)、堆囊菌(Sorangium)、气单胞菌(Aeromonas)、珊瑚球菌(Corallococcus)和地杆菌(Geobacter). 在湿地中还发现了其他细菌,如动杆菌(Proteobacteria)、绿弯菌(Chloroflexi)、硬壁菌门(Firmicutes)、酸杆菌(Acidobacteria)、拟杆菌门(Bacteroidetes)、硝化螺菌门(Nitrospirae)、放线菌(Actinobacteria)、螺旋菌门(Spirochaetae)、浮霉菌(Planctomycetes)、绿菌门(Chlorobi)、脱铁杆菌门(Deferribacteres)、蓝细菌(Cyanobacteria)、OP8,WS3,TA06和OP11,其中优势细菌为动杆菌(Proteobacteria)、绿弯菌门(Chloroflexi)和硬壁菌门(Firmicutes)[6]. 还有研究在湿地中发现了降解甲苯等污染物的细菌如皮氏罗尔斯顿菌(Ralstonia pickettii)[7].
光合细菌主要分为紫色非硫细菌、紫色硫细菌、绿色硫细菌和绿色丝状菌等4类. 光合细菌可以降解富营养水体中的污染物,是湿地发挥生态恢复功能的关键细菌. 在废水处理过程中,可进行光合作用而不产氧的光合细菌既可去除污染物,还可以通过生物炼制回收蛋白质、辅酶Q10、5-ALA、类胡萝卜素、细菌叶绿素和生物可降解塑料-聚羟基脂肪酸酯等物质和能量,是生态友好型微生物[8]. PSB对各营养型废水的化学需氧量、氨氮及总磷降解率可达到90%以上. 光合细菌还能去除养殖水体中氨氮和亚硝态氮. 光合紫色非硫细菌沼泽红假单胞菌(Rhodopseudomonas palustris)可以作为水质净化的添加剂[9]. 外硫红螺菌属(Ectothiorhodospira) 以单硫代砷酸盐为电子供体并将其完全转化为砷酸盐,参与硫和碳的生物地球化学循环[10]. 光合细菌还可以利用光能进行生物脱盐[11]. 类球红杆菌(Rhodobacter sphaeroides)可以用作生物监测和生物修复,去除放射性污染和汞污染[12]. 为了防止光照产生活性氧和紫外损伤,光合细菌光合基因的表达受到独特的调控. 其中包括调控RNA和反义RNA,反式作用的小非编码RNA[13]等.
但是,目前未见湿地光合细菌的研究报道,也未见生活污水对湿地光合细菌群落组成和多样性的影响的研究. 研究湿地光合细菌群落及其变化,将有助于丰富对湿地细菌区系及其功能的认识. 光反应中心复合体的M亚基的编码基因pufM是目前研究光合细菌的主要标记基因. 单拷贝的pufM基因广泛用在光合细菌的鉴定[14-22]或者定量[23]. pufM用作光合细菌生态研究的一般流程是扩增样品的pufM基因,构建文库、测序. 本研究第一次揭示了湿地光合细菌种类、群落结构,以及生活污水排放的影响,为认识湿地中光合细菌群落结构、生态学作用,以及关键功能菌的富集培养、分离和后续的基因组、生物化学、菌种之间相互作用提供了基础.
HTML
-
采集重庆北碚马鞍溪湿地公园淤泥样品(上游T1608A_B28和下游T1608A_B27,北纬29°49′33″,东经106°25′0″,上下游距离300 m,排污口位于上下游中间,距离上游50 m处),装入含DNA保护液的采集管. 样品用MIO-BIO PowerSoil DNA Isolation Kit抽提基因组DNA. 样品一式3份.
-
文库构建采用两步PCR扩增的方法,首先采用特异引物(即内侧引物)扩增目的片段,pufM基因引物序列F 5′-TACGGSAACCTGTWCTAC-3′,R 5′-AYNGCRAACCACCANGCCCA-3′. 胶回收目的片段作为模板进行二次PCR扩增(即外侧引物),将illumina平台测序所需的接头,测序引物,barcode添加到目的片段的两端.
引物如下:
F内侧引物:5′-TTCCCTACACGACGCTCTTCCGATCT-特异引物-3′
F外侧引物:5′-AATGATACGGCGACCACCGAGATCTACAC-barcode-TCTTTCCCTACACGACGCTC-3′
R内侧引物:5′-GAGTTCCTTGGCACCCGAGAATTCCA-特异引物-3′
R外侧引物:5′-CAAGCAGAAGACGGCATACGAGAT-barcode-GTGACTGGAGTTCCTTGGCACCCGAGA-3′
第1次PCR扩增 PCR扩增体系:5xBuffer 10 μL,dNTP (10 mmol/L) 1 μL,Phusion超保真DNA聚合酶1U,F/R内侧引物(10 μmol/L) 各1 μL,模板5~50 ng,ddH2O补至50 μL. ABI9700 PCR仪PCR扩增程序:94 ℃ 2 min,94 ℃ 30 s,50 ℃ 30 s,72 ℃ 30 s,35个循环,72 ℃ 5 min,10 ℃保温. 取3 μL PCR产物于1.2%的琼脂糖凝胶电泳检测.
第2次PCR扩增 切取第1次PCR凝胶电泳产物,采用AXYGEN公司的AxyPrepDNA凝胶回收试剂盒回收. 回收产物进行第2次PCR扩增. 第2次PCR扩增的体系与第一次类似,只是循环为8次. 取3 μL PCR产物于1.2%的琼脂糖凝胶电泳检测.
文库构建 将PCR扩增产物于2%琼脂糖凝胶电泳切胶回收(电泳槽保持干净,更换电泳缓冲液). 采用AXYGEN公司的AxyPrepDNA凝胶回收试剂盒回收. 回收产物进行qPCR定量. FTC-3000TM real-time PCR仪实时荧光定量,样本按照等摩尔比混匀后完成文库制备.
-
微基生物科技(上海)有限公司Illumina MiSeq 2×300 bp多个样品混合测序. 根据标示样本来源的barcode标签序列区分、提取各个样品的数据,以fastq格式保存. 特异引物去除序列中可能含有的模糊碱基、单碱基高重复区以及PCR过程中产生嵌合体. 过滤拼接的长Reads中的singletons(对应reads只有一条的序列),进行聚类OTU(operational taxonomic units). 采用软件为mothur V.1.39.5.
-
将OTU代表序列与NCBI进行Blast比对,找到相似度最高的已知物种的信息,排除未培养的环境样本信息,将该物种的分类信息注释为对应OTU的物种分类信息,通过物种信息的登录号得到各个级别的分类信息.
-
将OTU综合分类表中的信息按门、纲、目、科、属、种6个分类水平提取,分别统计各样品在不同分类水平比对上的序列数.
-
序列用NCBI数据库手动去掉载体,应用BLASTn程序与数据库进行相似性比对搜索,选择同源性最高的3条序列为参比序列. 采用ClustalX 2软件进行序列多重比对,然后以Kimura-2模型计算进化距离,以邻接法和MEGA 4.0软件构建系统发育树.
-
通过MOTHUR (http://www.mothur.org/wiki/Main_Page)软件进行序列的操作OTU的聚类,以Cutoff=0.03为阈值划分OTU. 并用GenoDive软件(http://genodive.org/)计算出多样性相关指数:有效基因型数、基因型多态性指数、均匀度指数、香农-威纳指数、修正香农-威纳指数、未校正的基因型多样性指数. 覆盖率运用公式:Rcov=1-(N/nInd)计算,N为整个文库中只出现一次的克隆子数目;nInd为文库中筛选出来的总的阳性序列数目. 稀释曲线运用POST软件计算,Excel绘图.
1.1. 样品的材料和处理
1.2. PCR扩增及文库构建
1.3. 测序与系统发育分析
1.3.1. OTU聚类
1.3.2. 物种分类学统计
1.4. BLAST分析
1.5. 多样性指数和样品聚类分析
-
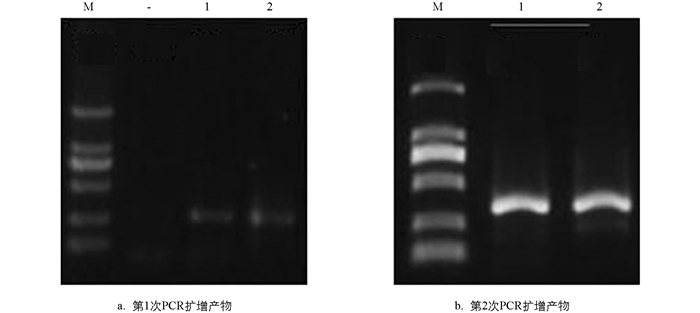
取第1次扩增的3 μL PCR产物于1.2%的琼脂糖凝胶电泳检测,结果见图 1a. 显示成功扩增pufM. 图 1b为第2次PCR扩增结果. 图中可见条带单一,亮度适中,可进行后续胶回收实验.
-
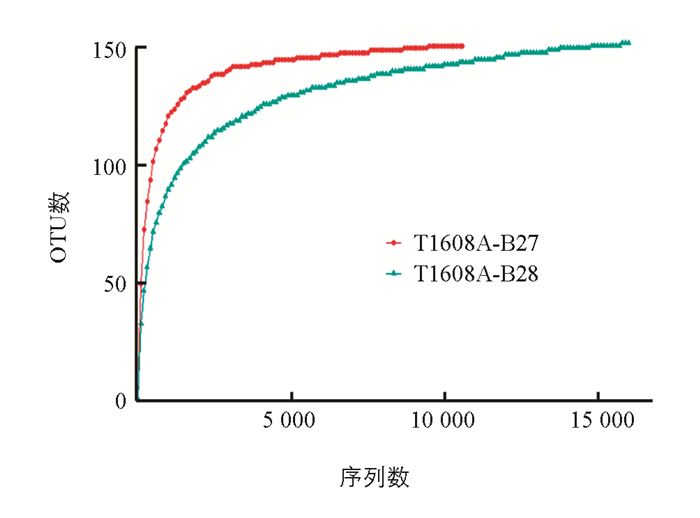
每个采样点采集3个样品,以下数据仅呈现代表性的结果. T1608A_B27和T1608A_B28两个样本的优化序列数分别为10,576和16,494. 为了确定样本的测序数据量是否合理,比较测序数据量不同的样本中物种的丰富度,从样本序列中随机抽样,以抽到的序列数与它们所能代表OTU的数目构建稀释性曲线. 当曲线趋向平坦时,说明测序数据量合理,更多的数据量只会产生少量新的OTU,反之则表明继续测序还可能产生较多新的OTU. 从稀释曲线(图 2)可看出光合细菌的测序数量合理,多样性采集较为充分.
-
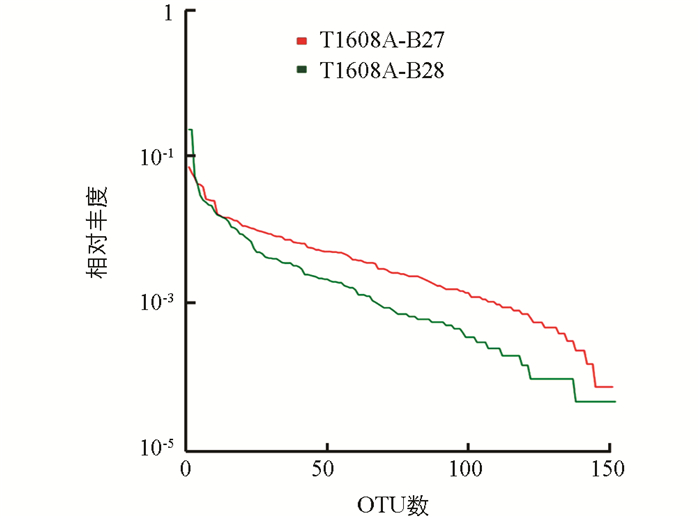
为了分析生活污水排放对光合细菌多样性的影响,统计两个采样点6份样品的结果. 下文描述其中代表性样品中每一个OTU所含的序列数,将OTUs按丰度(所含有的序列条数)由大到小的等级排序,再以OTU等级为横坐标,以每个OTU中所含的序列数为纵坐标作图,构建丰度等级图. 从图 3可见,生活污水排放影响马鞍溪湿地光合细菌的多样性,这体现在生活污水排放影响样品中光合细菌群落的丰富度,即群落中光合细菌的种类多少,不考虑群落中每种光合细菌的丰度. 这可以用chao指数和ace指数衡量,指数越大,说明样品中的物种越丰富.
图中水平方向,曲线的宽度反映物种的丰度,物种的丰度越高,曲线在横轴上的范围越大;曲线的形状(平滑程度)反映了样品中物种的均度,曲线越平缓,物种分布越均匀.
从表 1中可见,无生活污水排放的样品T1608A_B27中,光合细菌的种类较多,但进一步分析显示主要集中在低丰度的光合细菌. 生活污水排放降低了低丰度光合细菌的种类和数量.
生活污水排放影响群落的多样性. 群落多样性受样品群落中物种丰富度和物种均匀度的影响. 相同物种丰富度的情况下,群落中各物种均匀度越大,则群落多样性越大. 香农-威纳指数越大,说明物种越丰富. 从表中可见,无生活污水排放的T1608A_B27样品中,香农-威纳指数更大,提示光合细菌群落多样性更大. 而生活污水排放,降低了光合细菌的多样性. 辛普森指数越小,说明样品中的物种越丰富. 从表中可见,未受生活污水排放样品的辛普森指数更小,说明其中物种更丰富.
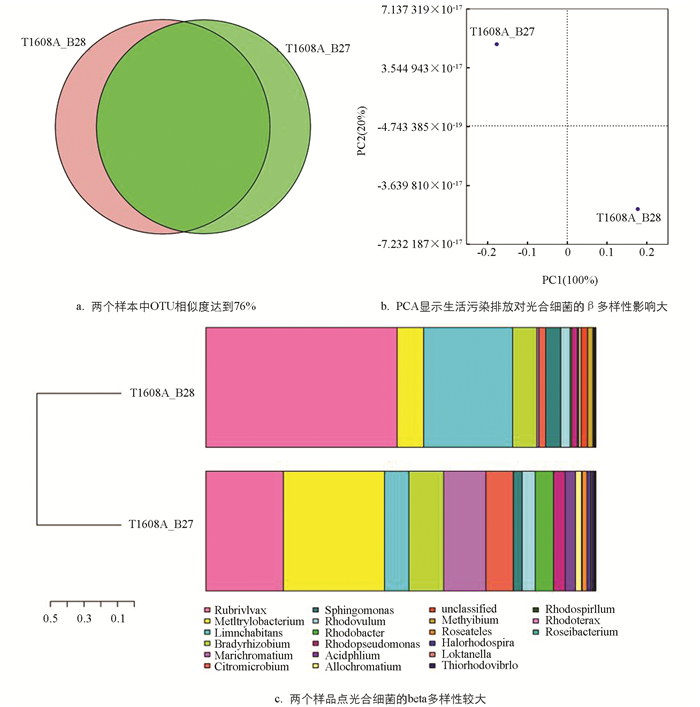
为了找出两个样点中相同的光合细菌,选用相似水平为97%的OTU样品,构建维思图. 从图 4a可见,两个样本中115个OTU相同,相似度达到76%,提示OTU组成非常相似.
为了分析两个样点间的差异和距离,我们分析两个代表性样品OTU(97%相似性)组成,进行主成分分析PCA(Principal Component Analysis). 根据每个样品OTU在每个样品的丰度文件计算出每个OTU在每个样品的比例,利用这个比例信息进行OTU的PCA分析. 从图 4b可见,生活污水排放对光合细菌的影响大.
生活污水排放对两个样点的光合细菌多样性影响很大. 图 4c显示生活污水排放后,光合细菌的beta多样性也明显降低,辛普森指数更高,提示β多样性降低.
-
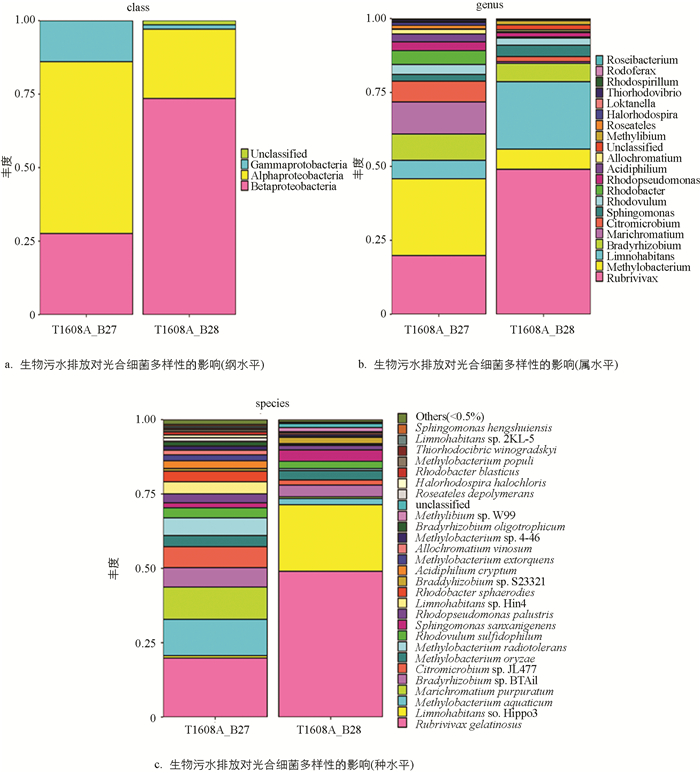
为了确定生活污水排放对光合细菌群落中哪些具体类群有显著影响,比较了两个样品中具体的微生物种类和相对丰度(序列数). 我们分别从纲、属和种3个水平进行了分析. 从图 5a可见,在纲水平上,组成都是α-变形菌、β-变形菌和γ-变形菌. 但是,生活污水排放处的β-变形菌是无生活污水的近3倍,α-变形菌和γ-变形菌在无生活污水排放点则是有污水排放的近3倍. 这提示生活污水排放显著改变纲水平的光合细菌群落结构. 从图 5b可见,在属水平上,生活污水排放后,明显增多的是红长命菌(Rubrivivax gelatinosus),湖栖菌(Limnohabitans sp. Hippo3),明显降低的是甲基杆菌(Methylobacterium aquaticum),海洋着色菌(Marichromatium purpuratum),柠檬色微菌(Citromicrobium sp. JL477)和红杆菌(Rhodobacter).
图 5c显示,生活污水排放后,在种水平上明显增多的是胶状红长命菌(Rubrivivax gelatinosus)和湖栖菌(Limnohabitans sp. Hippo3),明显降低的有水生甲基杆菌(Methylobacterium aquaticum)、紫色海洋着色菌(Marichromatium purpuratum)和柠檬色微菌(Citromicrobium sp. JL477).
2.1. 扩增获得了pufM片段
2.2. 两个样点的光合细菌测序深度合理
2.3. 生活污水排放影响光合细菌物种丰度和物种均匀度
2.4. 生活污水通过影响光合细菌群落中的少数种类而改变群落结构
-
为了探讨生活污水排放对湿地光合细菌群落组成和结构的影响,采用pufM标记基因,比较了湿地生活污水排放口上下游两个样点中光合细菌的差异. pufM基因是光合操纵子中的一个相对保守的基因,在亿万年的演化后依然同源性很高,常被作为光合细菌分子生态的标记基因.
研究结果发现的差异较大的几种光合细菌都报道过在污水处理中具有重要作用. 胶状红长命菌是紫色非硫细菌类光合细菌(non-sulfur photosynthetic bacterium),可以改善水质[24],是兼性光异养,生活在淡水环境如小池塘、食品加工废水、活性污泥和下水道中. 这与生活污水排放处该菌数量增加比较吻合. 根据环境条件,胶状红长命菌可以在浮游状态和生物膜状态下生长[25-26]. 同时,其代谢能力多样,可以光能营养,也可以化能营养,发酵,产氢(H2)[27],全基因组序列也包含了相应基因[28]. Mg2+[29]、Fe3+[30]均可提高胶状红长命菌生物量. 胶状红长命菌可以一氧化碳作为唯一碳源、水作为电子受体产生CO2和H2[31].
湖栖菌(Limnohabitans)属细菌是全球淡水生态系统中的优势菌[32],也是硫氧化菌,还参与氮循环,利用细菌叶绿素a捕获光能[33]进行光自养,是好氧不产氧光合细菌. 湖栖菌基因组具有光合基因簇、RuBisCO、一氧化碳脱氢酶、氨单加氧酶和硫氧化基因[32],提示代谢能力多样化. 营养会影响湖栖菌丰度[34-36]. 城市河流中,湖栖菌属细菌的丰度与溶解有机物的数量正相关[37]. 这与我们发现生活污水排放处其丰度显著增加也非常吻合. 根据营养条件不同,Limnohabitans属菌可能入侵,改变原有细菌区系生态,维持细菌多样性[38]. Limnohabitans属菌也是研究入侵物种对微生物区系影响的理想模式菌.
生活污水排放降低了水生甲基杆菌(Methylobacterium aquaticum)丰度. 镧系元素是甲基营养细菌XoxF-型甲醇脱氢酶的必需因子[39]. 与植物共栖的水生甲基杆菌基因组具有促进植物生长和利用植物释放的甲醇的基因[40],这提示该类菌可能与水生植物一起发挥作用.
紫色海洋着色菌(Marichromatium purpuratum)属于紫色硫细菌,其硫代硫酸脱氢酶含有两个血红素,属于c-型细胞色素[41],它们会积累元素硫[42].
柠檬色微菌(Citromicrobium)在寡营养的环境中广泛存在[43],也属于好氧不产氧光合细菌. 生活污水排放点其丰度降低,与其在寡营养环境中广泛存在吻合.
当然,本研究DNA水平的结果只能反映环境中光合细菌整体的多样性,不能反映出群落中光合细菌的生存能力、代谢活力以及生长力,也不知优势光合细菌pufM表达水平高低和功能. 下一步需要在转录水平以及生态功能水平研究湿地光合细菌,特别是分离这些光合细菌,比较它们处理生活污水的能力.





 DownLoad:
DownLoad: