-
开放科学(资源服务)标识码(OSID):

-
株高是水稻的重要农艺性状之一,也是构建理想株型的一个重要因素. 株高与水稻光合效率、抗倒伏以及产量有着密切的联系. 20世纪的绿色革命在矮化株型育种中取得了巨大成功,然培育兼顾产量和抗倒伏的适宜株高仍是水稻、玉米和小麦等大多数作物的首要育种目标,因此水稻株高的遗传及分子机制一直是研究的热点.
水稻株高主要由节间细胞的数目和大小决定. 节间细胞的增殖及扩张受多重因素的影响,包括植物激素、细胞周期、细胞壁合成、表观遗传学及转录因子等,同时也受到外界环境因素的影响,如光照、温度、物种间竞争等. 植物激素作为植物中重要的调节因子,参与植物的整个生长发育过程,其中赤霉素(GA)和油菜素内酯(BR)是水稻株高的重要决定因子[1-2]. SD1编码赤霉素生物合成的关键酶GA20ox,突变后导致GA20ox失活,造成株高降低[3-4]. EUI1编码细胞色素P450单加氧酶CYP714D1,eui1突变体及eui1 RNAi植株赤霉素含量升高,倒1节显著伸长,株高增加,而过表达植株中赤霉素含量降低,导致株高降低,因此EUI1负调控赤霉素介导的细胞伸长[5-6]. D35/OsKO2编码贝壳杉烯氧化酶,参与催化赤霉素生物合成的早期步骤,其功能缺失导致d35株高严重降低[7]. SLR1编码1个DELLA蛋白,是赤霉素信号传导的负调控因子,其功能是导致突变体株高降低. GID1编码1个可溶的赤霉素受体,类似于激素敏感脂肪酶,并且对具有生物活性的GAs具有高度的亲和性,GID1过量表达表现出GA高敏感性表型. GID1与SLR1相互作用,从而形成GID1-GA-SLR1复合体,阻止赤霉素信号向下传递[8-11].
油菜素内酯能够促进植物生长,在细胞伸长及分裂中也具有重要作用. BRD1,BRD2和D2均参与油菜素内酯的生物合成,分别编码C-6氧化酶、氧化还原酶和细胞色素P450,这些基因的突变导致油菜素内酯生物合成受阻,突变体中油菜素内酯含量降低,表现出严重矮化[12-14]. BRI1和BZR1参与油菜素内酯信号转导,分别编码油菜素内酯受体激酶和下游信号分子. bri1第2节不伸长,株高矮化,并且对外源油菜素内酯不敏感;BZR1能够调控下游油菜素内酯响应基因,干涉植株表现为矮杆[15-16].
过氧化物酶是一大类催化各种底物发生氧化的生物体保护酶类家族,在植物的生长发育中具有重要作用. 根据功能不同,将过氧化物酶分为3类:Ⅰ类(抗坏血酸)过氧化物酶、Ⅱ类(木质素)过氧化物酶和Ⅲ类(分泌型)过氧化物酶[17]. Ⅲ类过氧化物酶(CⅢ PRXs)广泛分布于植物界并且是植物中特有的一类过氧化物酶,参与植物激素分解代谢、木质素生物合成、种子萌发、细胞伸长和防御病原体等多种生物生理过程. CⅢ PRXs基因编码含血红素的酶,接受来自不同分子供体的电子,催化H2O2的代谢,产生活性氧(ROS,即OH-或O2-). 这些特性使得CⅢ PRXs能够在过氧化循环过程中调控损伤植物细胞的ROS产生[18]. CⅢ PRXs酶活性由ROS触发,ROS的产生由不同的环境刺激诱导,以抵御潜在的威胁[19].
Ⅲ类PRX家族较为庞大,在水稻和拟南芥中分别包含138个和73个成员,其中大部分的生物学功能并不清楚[20-21]. 拟南芥中研究表明过氧化物酶与植物生长呈负调控,AtPRX71通过在外质体中积累H2O2抑制细胞扩张,atprx71突变体莲座叶变大、生物量显著增加,而过表达植株生长缓慢莲座叶变小、生物量显著下降,并且AtPRX71表达水平的升高会促进ROS的积累导致细胞壁成分变化[22]. AtPRX37过表达植株生长迟缓,表现为矮化[23],因此AtPRX71和AtPRX37均是拟南芥生长的负调控因子. AtPRX72,AtPRX52和AtPRX4参与拟南芥木质素的生物合成,atprx72生长缓慢,叶片形态及角果数减少等. AtPRX52参与木质化过程中微管纤维的合成,突变体中木质素含量降低,木质素生物合成相关基因的表达下调[24-25]. OsPRX38在拟南芥中过表达后激活不同抗氧化系统信号网络,促进木质素生物合成,增强根部的木质化程度,导致过表达植株中砷积累减少[26]. AtPRX17调控木质化组织的形成,在营养生长向生殖生长的转变过程中受到AGL15的调控[27]. 在水稻中,OsPRX30通过调节过氧化物酶体(POD)的活性与ROS的含量来调节白叶枯病抗性,而OsPRX30的表达又受到含有AT-hook结构域的转录调节因子OsATH1的调控,进一步的研究揭示了OsATH1通过调节OsPRX30启动子区内的组蛋白H3乙酰化水平进而影响OsPRX30表达以调控白叶枯病抗性[28]. 目前为止,仅有少数Ⅲ类PRX基因生物学功能被揭示,还有大部分尚未报道,尤其在水稻中还未见Ⅲ类PRX基因参与调控株高的相关报道.
本研究通过CRISPR/Cas9基因编辑技术,在粳稻中花11背景下创制的Ⅲ类过氧化物酶家族敲除突变体库中鉴定到1个株高变高的突变体,将其命名为iph1(increaseing plant height 1). 与野生型相比,iph1的节明显变长,导致株高增加. 本研究通过表型分析、农艺性状考察、表达模式分析、过氧化物酶活性分析、亚细胞定位等对IPH1进行功能分析,探究IPH1在调控水稻株高发育及株型形态建成中的作用,为利用该基因进行水稻株型分子设计育种奠定基础.
HTML
-
水稻突变体iph1来源于以中花11为背景的敲除突变体库,经过多代自交,性状能够稳定遗传. 所有试验材料均种植于西南大学水稻研究所.
-
通过NCBI数据库的Blast工具获取IPH1的结构域信息和同源蛋白序列,下载同源蛋白序列,并将其保存为FASTA文件格式,将FASTA文件导入MEGA5软件,根据贝叶斯最低分准则利用Neighbor-joining方法构建进化树,对IPH1进行进化分析.
-
利用天根RNA提取试剂盒(TIANGEN生物公司)提取野生型和iph1突变体总RNA,并用宝生物PrimeScript试剂盒(TaKaRa)将RNA反转录成cDNA. 以水稻ACTIN为内参基因,使用PrimerPrimer5.0软件设计引物(表 1)[28],用Bio-Rad实时荧光定量PCR仪(CFX Connect Realtime Reaction System)进行扩增,每个样品设置3个重复. 基因相对表达量计算用2-ΔΔCt法,并用t检验进行差异显著性分析.
-
为了确定IPH1的亚细胞定位,使用引物pAN580-IPH1-F和pAN580-IPH1-R(表 1)从野生型中扩增不含终止密码子的IPH1编码序列,将该片段克隆到瞬时表达载体pAN580的XbalⅠ/Bam HⅠ位点上,构建IPH1-GFP载体. 经序列分析验证后,通过PEG介导法将pAN580-GFP和IPH1-GFP质粒转化水稻原生质体. 28 ℃孵育12~16 h后,用激光共聚焦显微镜(LSM800,Zeiss,德国蔡司)观察荧光信号并拍照.
-
取8周龄水稻幼苗的新鲜叶片(300 mg),在液氮中研磨成粉,并与4 mL 0.05 mol/L PBS(pH值为7.8)混合在5 mL试管中. 快速解冻后,将试管7 500 r/min离心15 min,收集上清液(含POD). 以愈创木酚为供氢体,采用分光光度计在405 nm处测定OD值,根据公式计算过氧化物酶活性[29].
-
将野生型和iph1突变体新鲜组织于液氮中充分研磨,加入9倍体积的生理盐水,冰水浴条件下机械匀浆,1 200 r/min离心10 min,取上清液待测. 每个样本设置3个重复,采用南京建成生物工程研究所的H2O2试剂盒测定H2O2质量分数.
1.1. 试验材料
1.2. 生物信息学分析
1.3. RNA提取及RT-qPCR分析
1.4. 亚细胞定位
1.5. 过氧化物酶活性分析
1.6. 过氧化氢(H2O2)质量分数测定
-
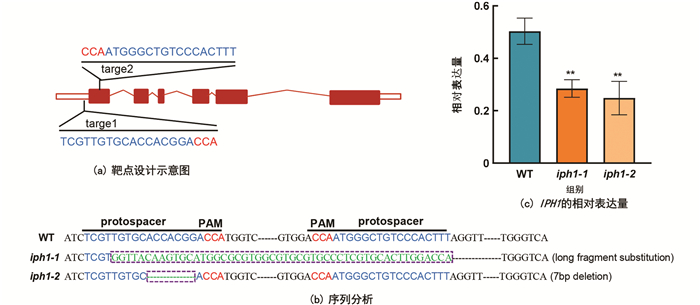
CRISPR/Cas9表达载体构建的靶点选择如图 1a,在基因(LOC_Os12g09460)的5′-UTR及编码区前端设计了2个靶点. 通过农杆菌介导转化中花11愈伤,对转基因植株靶点测序,比对发现2个独立的转基因株系在PAM(protospacer adjacent motif)附近发生了2种方式的编辑:iph1-1靶点1处发生了大片段替换,导致IPH1蛋白翻译的起始密码子缺失,无法正常翻译;iph1-2在目标位置缺失了7个碱基(ACCACGG)(图 1b). 进一步通过qRT-PCR分析,IPH1在两种编辑方式的突变体中表达均极显著下调(图 1c). 以上结果表明所构建的iph1突变材料为过氧化物酶基因(LOC_Os12g09460)敲除突变体,可用于后续的表型及生理生化等分析.
-
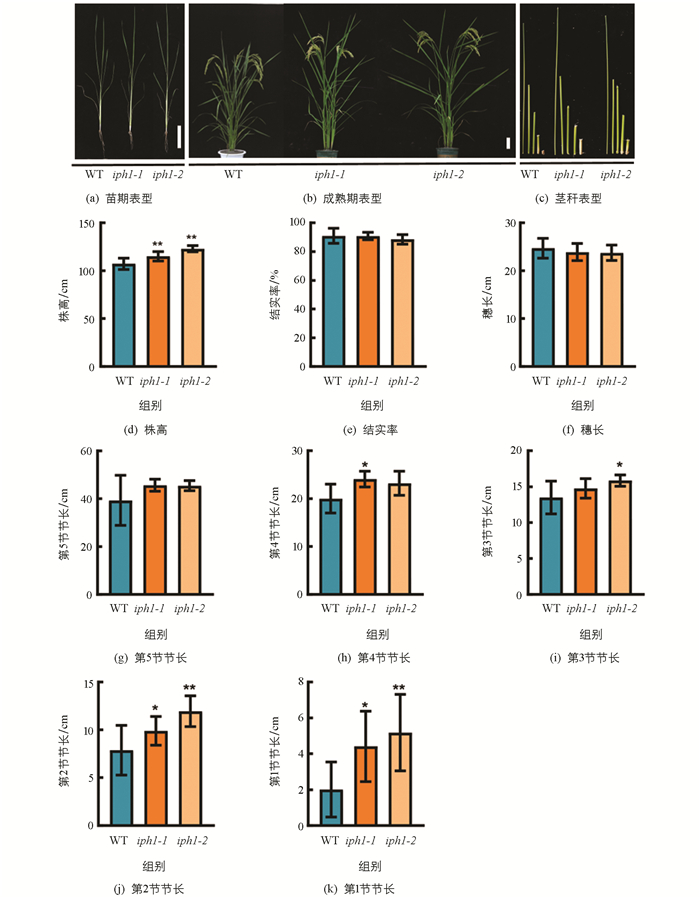
对iph1突变体植株经过多代全生育期的田间观察和统计,发现iph1突变株在苗期较野生型差异无统计学意义(图 2a),在抽穗期至成熟期阶段株高出现差异,成熟期iph1突变体的株高显著高于野生型(图 2b),iph1-1和iph1-2的株高分别增加了7.4%,14.6%(图 2d). 结果表明IPH1基因功能缺失主要影响株高,且遗传性状稳定.
成熟期时,对野生型和iph1-1,iph1-2突变体的结实率、穗长和各节间长度进行了详细统计. 结果发现,iph1-1,iph1-2突变体的结实率、穗长以及倒1节(即第5节)节长较野生型差异无统计学意义(图 2e至2g),iph1-1突变体的第4节节长较野生型显著增加,iph1-2突变体的第3节节长较野生型显著增加,iph1-1和iph1-2突变体位于茎基部的第1节和第2节节长较野生型均显著增加(图 2c,2h至2k). 该结果表明突变体株高变高主要由茎基部节长变化所致.
-
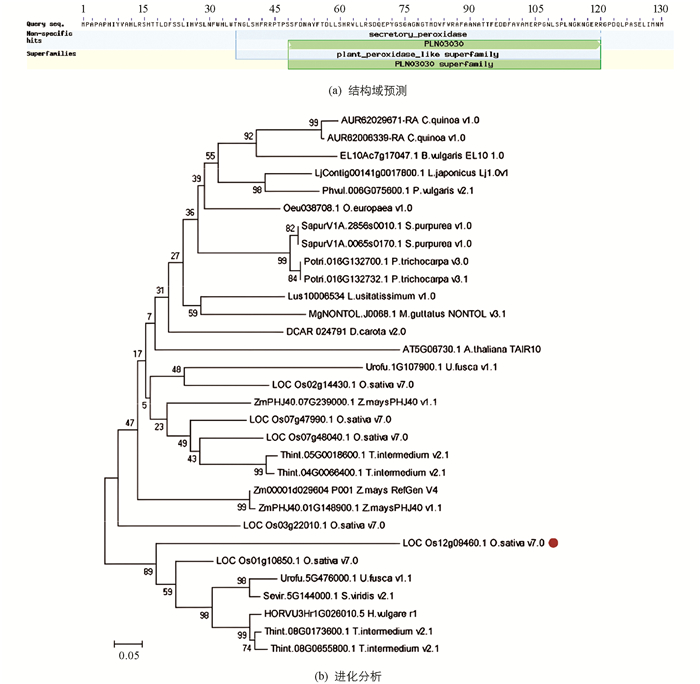
通过NCBI数据库获取IPH1的结构域信息,发现IPH1是一个过氧化物酶基因,在氨基酸第36~120区间内含有1个分泌型过氧化物酶(secretory peroxidase)结构域,属于第Ⅲ类过氧化物酶家族(图 3a);同时也获得了IPH1蛋白在其他物种的同源蛋白序列,将其保存为FASTA文件格式,根据贝叶斯最低分准则,利用MEGA5软件的Neighbor-joining方法构建进化树,分析发现IPH1编码的蛋白独立于一个分支,具有高度保守性(图 3b).
-
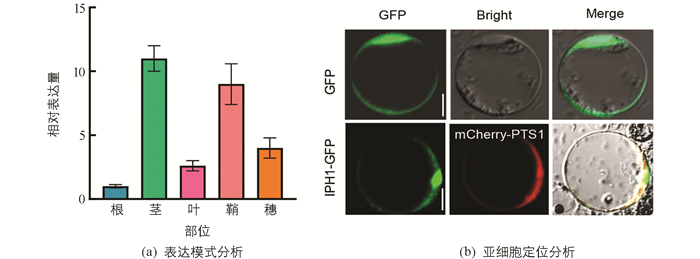
为了确定IPH1基因在水稻各组织中的表达情况,我们以野生型根、茎、叶、鞘和穗的cDNA作为模板,进行实时荧光定量表达分析. 结果显示IPH1转录本在不同部位均有表达(图 4a),其中在茎和鞘中有较高表达.
IPH1蛋白属于第Ⅲ类过氧化物酶家族蛋白,为了探究IPH1在细胞内部细胞器的定位情况,将CaMV35S-IPH1-GFP融合蛋白及过氧化物酶体CaMV35S-PTS1-mCherry融合蛋白共同转化水稻原生质体,同时将转化含CaMV35S-GFP的空载体pAN580作为对照,28 ℃避光过夜培养18 h,利用激光共聚焦显微镜观察荧光信号. 结果发现大部分绿色荧光都能与红色荧光重叠(图 4b),表明IPH1主要定位于过氧化物酶体中.
-
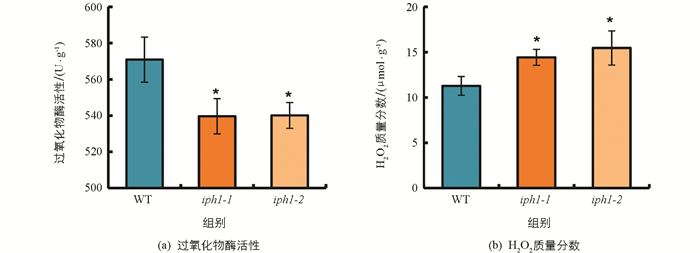
IPH1编码1个Ⅲ类过氧化物酶,但IPH1是否影响过氧化物酶的活性和H2O2质量分数尚不清楚,因此,采用分光光度法测定野生型及iph1突变体植株中的过氧化物酶活性和H2O2质量分数. 结果显示,iph1突变体的过氧化物酶活性显著降低,H2O2质量分数显著增加(图 5).
2.1. iph1敲除突变体的获得及鉴定
2.2. 表型鉴定
2.3. 生物信息学分析
2.4. 表达模式及亚细胞定位分析
2.5. 过氧化物酶活性及H2O2质量分数分析
-
IPH1是位于水稻第12染色体上的一个新基因,目前还未见其生物学功能的相关报道. 蛋白序列分析发现IPH1包含1个分泌型过氧化物酶结构域,属于第Ⅲ类过氧化物酶(Class Ⅲ peroxidases,CⅢ PRXs)家族. CⅢ PRXs作为一类植物特异性氧化还原酶参与植物激素的分解代谢、木质素的生物合成、细胞伸长和病原菌防御等多种生物学过程[18],水稻和拟南芥中CⅢ PRXs家族成员分别有138个和73个[21]. 目前在水稻中对CⅢ PRXs家族基因的功能研究较少,仅有1个家族基因OsPRX30被报道. 该基因编码1个主要定位于内质网的过氧化物酶前体蛋白,对细菌性黄单胞菌有反应,过表达OsPRX30会维持高水平的过氧化物酶(POD)活性和降低过氧化氢质量分数,从而增强植物对白叶枯病的敏感性. 进一步研究揭示含有AT-hook结构域的转录因子OsATH1通过调节OsPRX30启动子区内的组蛋白H3乙酰化水平进而调节OsPRX30表达,通过调控过氧化物酶体活性与ROS量调控白叶枯病抗性[30]. 本研究中,IPH1蛋白主要定位于过氧化物酶体内,在根、茎、叶、鞘、穗中均有表达,且在茎和鞘中表达相对较高.
-
水稻株高是非常重要的一个农艺性状,对抗倒伏及生物量的产生具有重要意义. 株高受到植物激素、细胞周期、细胞壁合成、表观遗传学、转录因子以及光照、温度等多重因素的影响. 如SD1,EUI1,D35/OsKO2,SLR1,GID1等通过调控赤霉素的合成及信号传导等调控细胞分裂或细胞伸长[3-6, 8-9]. BRD1,BRD2,D2,BRI1和BZR1通过参与油菜素内酯的生物合成及信号转导调控水稻株高[12-16]. 此外,一些基因的异常表达也会影响株高,OsCD1,BC12,OsGLP1等影响水稻细胞伸长或分裂的基因异常表达,导致产生矮化或半矮化表型[31-33]. 本研究中,鉴定到1个编码Ⅲ类过氧化物酶的基因IPH1影响水稻株高. 有关CⅢ PRXs调节植物株高的报道相对有限,小麦新型矮杆突变体Rht-B13b中CⅢ PRXs在扩张组织中表达显著上调,CⅢ PRXs可以使用H2O2增加细胞壁交联,从而导致细胞扩增和生长减少,最终使得植株表现为矮化表型[34]. 本研究中,CⅢ PRXs家族基因IPH1不同方式的突变均导致植株高度增加;进一步研究发现,iph1突变体中过氧化物酶活性降低,而H2O2的质量分数显著增加,因此IPH1可能通过过氧化氢途径调控水稻株高的形成,但是具体的分子作用机理仍需进一步研究.
3.1. IPH1编码1个含有分泌型过氧化物酶结构域的新基因
3.2. IPH1可能通过过氧化氢途径调控水稻株高
-
本研究鉴定了1个在粳稻中花11背景下敲除的第Ⅲ类过氧化物酶家族突变体iph1. 与野生型相比,两种突变方式的iph1突变体株高变高的表型能够稳定遗传;进一步统计也发现iph1突变体的第1节和第2节节长较野生型显著增加. 生物信息学分析表明,IPH1基因包含1个分泌型过氧化物酶结构域,利用同源蛋白序列进行进化树分析,结果显示IPH1独立于1个分枝,具有高度保守性. IPH1基因在各个组织中均有表达且在茎秆中表达相对较高,可能基因在表达水平上会影响表型,亚细胞定位表明该蛋白主要定位于过氧化物酶体中. 生理分析发现突变体过氧化物酶活性降低,H2O2质量分数增加. 本研究为深入了解水稻株高遗传调控网络提供了基因资源,并进一步丰富了株高相关的生物育种材料.





 DownLoad:
DownLoad: