-
生长素是植物生长发育过程中重要的激素之一,在植物生命周期中发挥重要的调控作用,如参与植物的器官发生和形态建成、衰老、顶端优势及组织分化等[1-2].与生长素信号转导相关的3类主要蛋白组分即生长素/吲哚乙酸蛋白(auxin/indoleacetis acids proteins,Aux/IAAs)、生长素响应因子(auxin response factors,ARFs)和SCF复合体[3].其中Aux/IAA基因家族是生长素信号转导途径中重要的作用因子,在生长素的作用下Aux/IAA泛素化降解,Aux/IAA蛋白对ARF的抑制作用也随之解除,从而启动生长素信号途径.
目前,Aux/IAA基因家族已在拟南芥(Arabidopsis thaliana)、水稻(Oryza sativa)、杨树(Populus trichocarpa)、玉米(Zea mays)、高粱(Sorghum vulgare)、番茄(Solanum lycopersicum)、苹果(Malus domestica)、黄瓜(Cucumis sativus)、粗山羊草(Aegilops tauschii)和马铃薯(Solanum tuberosum)等[4-13]多个物种中进行了详细分析,研究表明该基因家族在植物果实发育中扮演着十分重要的作用.
在实际生产中,施用外源赤霉素能够诱导植物单性结实[14-16].枇杷果实为假果,果肉主要由花托发育而来.三倍体枇杷由于配子母细胞减数分裂异常,难以形成正常的花粉,造成花粉败育而不能正常授粉受精,其自然结实率极低,在施用外源赤霉素后坐果率明显升高.研究表明,生长素和赤霉素是促进坐果和果实生长发育的主要激素,生长素主要是促进细胞分裂,赤霉素促进细胞伸长.而Aux/IAAs基因是生长素精确反馈调节的关键环节,因此探讨赤霉素处理后生长素响应基因的变化对于果实坐果具有重要的意义.
本文以三倍体枇杷A322为研究对象,研究GA3处理后枇杷坐果期4种内源激素的变化,克隆了6个Aux/IAA基因家族的cDNA片段,进一步分析了这些基因在三倍体坐果期的表达情况,以期为三倍体无籽枇杷坐果机制研究提供理论参考.
全文HTML
-
以南方山地园艺学教育部重点实验室暨重庆市枇杷工程技术中心选育的三倍体枇杷A322为材料,材料采集于2017年10月下旬至2017年12月中旬.以花瓣未绽开的花朵为0 d,将其去雄后用GA3 50 mg/L[17]处理,以清水处理为对照.在处理后第1,3,5,7,14和21 d取下整朵单花或幼果,并将其剥为子房和花萼等其他部分,液氮速冻处理后,于-80 ℃保存备用.
-
内源激素含量采用高效液相色谱(HPLC)技术测定.内源激素的提取和测定均参照李华等[18]的方法进行.
-
参照艾得莱公司试剂盒说明书提取RNA并参照博日科技公司反转录试剂盒说明书进行反转录反应.
-
参照枇杷转录组测序中筛选出的Aux/IAAs基因,用软件Primer Premier 5.0设计引物(表 1). PCR反应体系总体积为25 μL:2×Taq PCR MasterMIX(12.5 μL),10 mmol/L Forward Primer (1 μL),10 mmol/L Reverse Primer(1 μL),1 μL cDNA模板,无菌去离子水(补足至25 μL). PCR反应扩增程序:94 ℃预变性3 min;94 ℃变性30 s,60 ℃退火30 s,最后72 ℃延伸1 min.
-
回收目的片段与pMD19-T载体在16 ℃下连接3~4 h.连接产物转化到大肠杆菌(E.coli) DH5α细胞中,经Amp抗性筛选和菌落PCR筛选,取阳性克隆送上海英潍捷基公司进行测序.测序结果在GenBank数据库进行BLAST序列比对分析.
-
以枇杷Actin为内参基因[19]5′-ATCCTTCGTCTGGACCTTGC-3′和5′-GACAATTTCCCGTTCAGCAGT-3′,根据扩增所得基因的CDS设计实时荧光定量引物(表 2).采用2-ΔΔCt法在ABI上进行实时荧光定量PCR,PCR反应设置3次重复.反应体系为10 μL:模板cDNA 2 μL,上、下游引物各0.2 μL,2×ChamQ SYBR qPCR Master Mix 5.0 μL,50×ROX Refence Dye 0.2 μL,ddH2O 2.4 μL. PCR反应程序为:95 ℃预变性30 s;95 ℃ 15 s,58~62 ℃ 30 s,40个循环后作熔解曲线.
-
试验数据用软件Excel 2007和Originpro 9.0进行分析和处理.
1.1. 试验材料
1.2. 内源激素的分离、提取和测定
1.3. 总RNA的提取及反转录
1.4. PCR引物设计及反应体系和扩增程序
1.5. PCR产物回收、阳性克隆筛选及测序
1.6. Real-time PCR分析
1.7. 数据处理
-
以三倍体枇杷花为材料,等量分成2份,其中1份加入标准品,另1份不加标准品,按照同样的流程完成激素提取后计算各激素的回收率.将4种激素标准品分别稀释成一系列浓度梯度,在相同的色谱条件下测定峰面积Y.以质量浓度X(μg/mL)为横坐标,色谱峰面积为纵坐标绘制标准曲线,计算相应的线性回归方程及相关系数,结果如表 3所示.各种激素线性回归方程的相关系数均在0.99以上,说明标准曲线可以用于样品中内源激素浓度的计算.回收率结果表明:IAA,GA3和ZT的回收率相对较高,ABA的回收率相对较低,但可满足后续分析的需要.
-
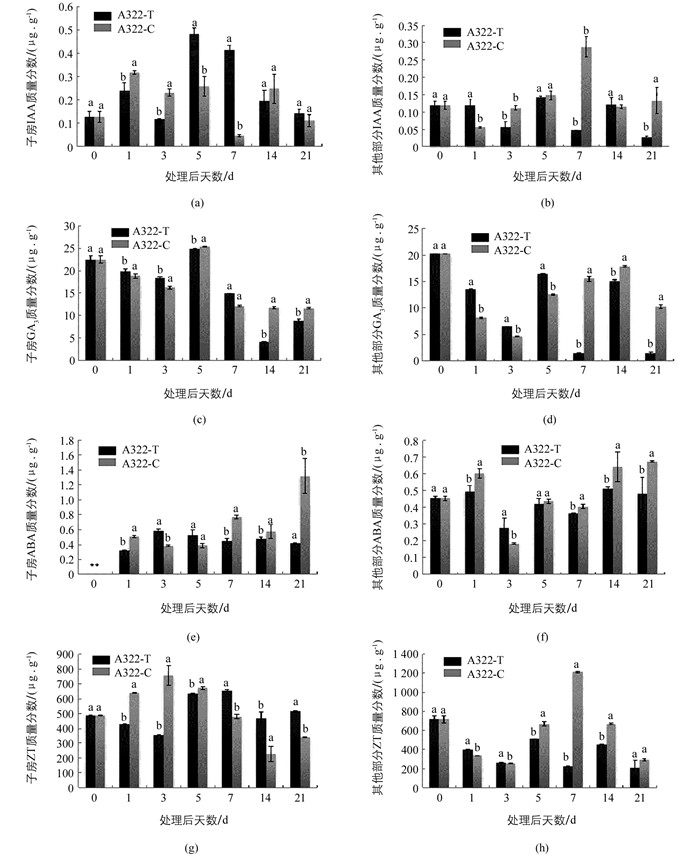
对三倍体枇杷A322坐果期子房及其他部分的内源激素测定结果表明:对于IAA而言,外源GA3处理5 d和7 d后,内源IAA出现2个高峰随后开始下降,而对照组子房内源IAA质量分数在清水处理7 d后最低,只有0.048 2 μg/g,约是处理组的1/10,推测子房内源IAA质量分数变化可能是枇杷坐果的关键因素之一.子房中,外源GA3处理后,ABA在处理21 d内,整体质量分数都较低.清水处理条件下,子房内源ABA在21 d达到高峰值,并显著高于处理组.对于其他部分而言,内源ABA质量分数除处理后第3 d外,其余天数对照组内源ABA质量分数均高于处理组.子房中,外源GA3处理后,除处理后第5 d外,内源GA3质量分数在处理当天最高.其他部分清水处理后第7 d开始,内源GA3质量分数显著高于处理组GA3的质量分数.对于ZT而言,子房中,外源GA3处理7 d后,处理组ZT质量分数显著高于对照组.其他部分除了对照处理第7 d检测到浓度较高外,其余天数内源ZT质量分数都低于处理当天(图 1).
-
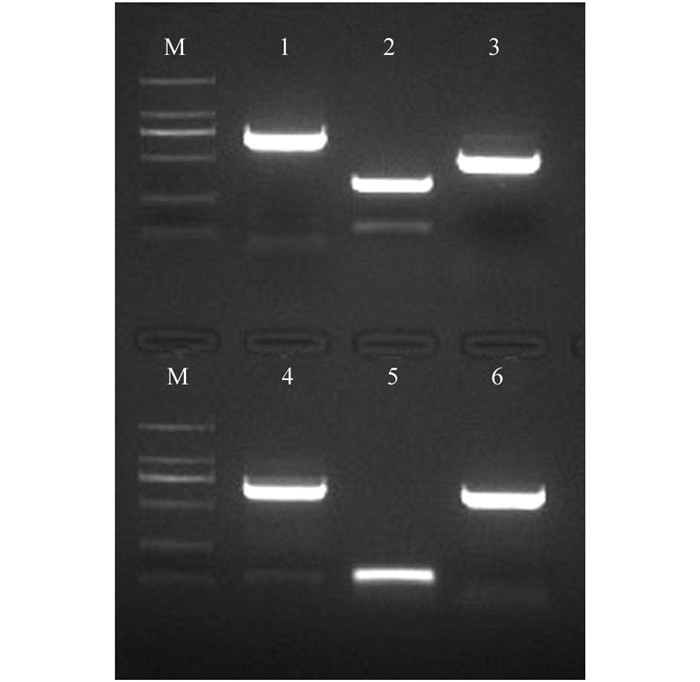
从前期转录组测序结果中筛选出6个Aux/IAA家族基因,以枇杷A322开花当天花朵cDNA为模板进行PCR扩增,获得的6个基因的cDNA片段(图 2).用BLAST进行核苷酸相似性检索,发现IAA6与苹果MdIAA6(HM122447.1)相似性为93%;IAA8与苹果MdIAA8(NM_001294015.1)相似性为97%;IAA9与苹果MdIAA9(NM_001293892.1)相似性为93%;IAA10与苹果MdIAA10(HM122440.1)相似性为90%;IAA11与苹果MdIAA11(NM_001293857.1)相似性为97%;IAA20与苹果MdIAA20(NM_001293857)相似性为95%;说明克隆的6个基因与预期结果一致.
-
以A322花或幼果的子房和其他部分为研究材料,以花瓣绽开前未经处理花的子房和其他部分各基因的表达量作为对照,分析这6个基因的表达特点.
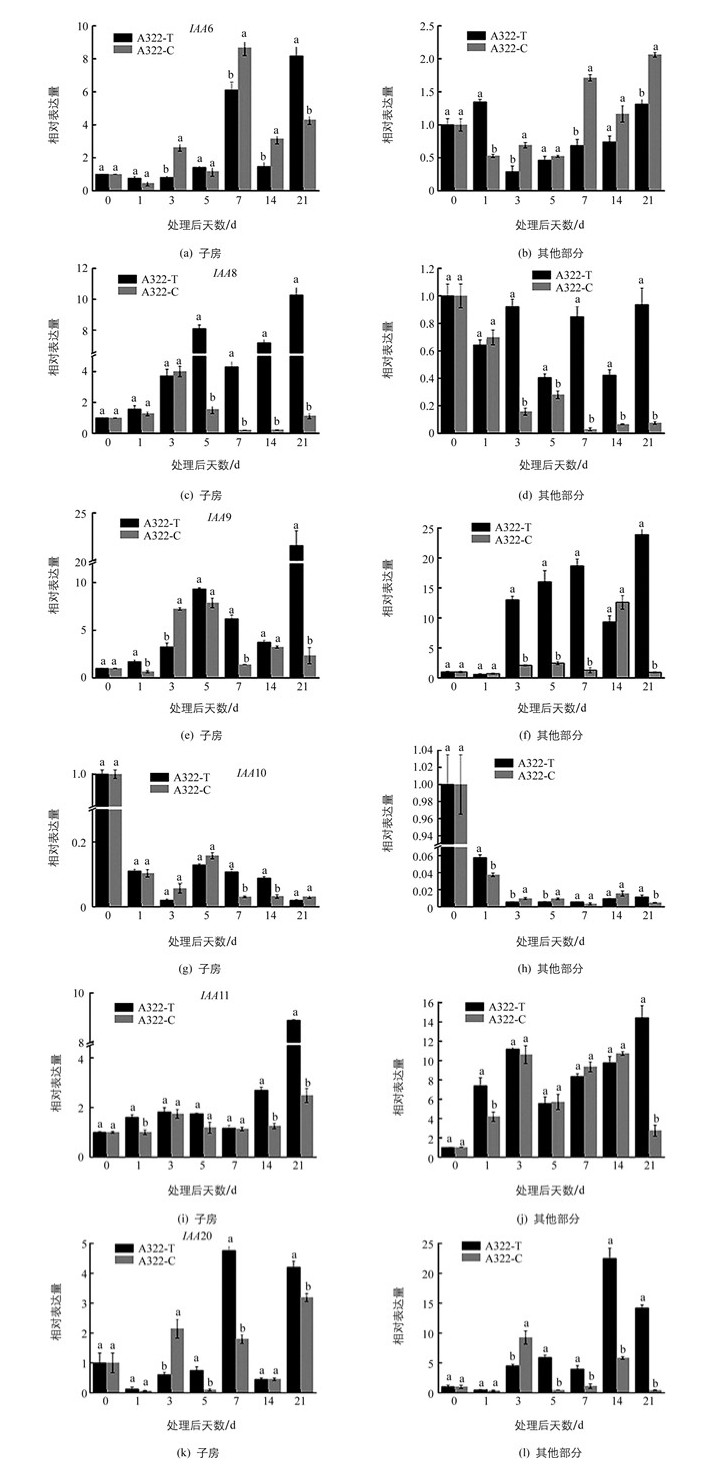
在已克隆和定量分析的Aux/IAA相关基因中,IAA6,IAA8,IAA10在子房中的表达量明显高于其他部分;IAA9在子房和其他部分的表达量相当,IAA11和IAA20在子房中的表达量低于其他部分.子房中,外源GA3处理后IAA6的表达量在21 d达到最大值.其他部分中,在处理3 d后,处理组IAA6表达量低于对照组.子房中,IAA8在GA3处理后表达量呈先上升后下降再上升的趋势,并在处理21 d后表达量最高.在前3 d处理组和对照组IAA8表达量相差不大,但在处理5 d后处理组表达量显著高于对照组.对于其他部分而言,IAA8在露白当天表达量最高,处理后无论是处理组还是对照组表达量均低于露白当天.对于IAA9而言,子房中,外源GA3处理后IAA9的表达呈先升高后降低、再升高的趋势,并在处理21 d后表达量达到最高值.清水处理1 d后,IAA9的表达出现了极低值,并在处理5 d后对照组IAA9的表达量一直低于处理组.对于其他部分而言,外源GA3处理3 d后基因IAA9的表达量一直保持在较高水平,并在21 d达到表达量最大值,对照组在清水处理14 d前表达量一直处于较低水平,且显著低于处理组的表达量.对于IAA10而言,表达量无论是在子房还是在其他部分,IAA10均在处理当天表达量最高(图 3).
2.1. 三倍体枇杷坐果期激素的测定
2.1.1. 检测方法的线性范围及回收率
2.1.2. 枇杷坐果期子房和其他部分内源激素的变化
2.2. Aux/IAA家族部分基因的克隆及同源性分析
2.3. Aux/IAA家族基因在枇杷坐果期不同组织表达特性分析
-
生长素在植物体整个生命周期中具有重要作用,在果实发育过程中,生长素可以调控果实坐果、成熟以及脱落[20].对于非单性结实类型植物而言,种子是产生IAA的主要部位.大量研究表明,外源GA3处理后子房内IAA质量分数增加[21-22].本研究检测坐果期子房和其他部分4种内源激素含量发现,外源GA3处理后,子房内IAA质量分数整体比其他部分高,且在GA3处理后,子房内出现2个IAA质量分数高峰值,且在第7 d处理组IAA质量分数约是对照组的10倍,推测外源GA3的应用促进了枇杷子房内源IAA升高,外源GA3提高枇杷坐果率可能部分通过影响内源IAA质量分数而起作用的. ABA是一种抑制生长的植物激素,高浓度ABA会促进果实脱落[23].本研究发现在处理前期ABA质量分数较低,子房中,清水处理7 d后对照组ABA质量分数开始升高,且显著高于处理组,并在21 d达到高峰,与田间观察的对照组落花动态时间相似.推测清水处理21 d后开始出现大量落花的原因可能与内源ABA质量分数升高有关.
Aux/IAAs是一类半衰期较短的核蛋白,普遍包含4个保守的结构域,在果实发育过程中有重要的调控作用[24].本研究发现,子房中,外源GA3处理5 d后IAA9的表达量上升,还发现在外源GA3处理前期,子房中IAA9的表达量与内源IAA含量变化趋势一致,推测IAA9的表达上调可能受到了内源IAA的诱导.葡萄中,VvIAA9是一个正调控因子,在葡萄开花到成熟过程中表达量都上调,拟南芥中超表达VvIAA9使拟南芥快速生长并提高植株对生长素的敏感性[25].番茄果实发育过程中,IAA9是一个负调控因子,沉默IAA9使番茄在未授粉之前就出现单性结实现象[26-27].本研究结果中IAA9的表达变化与葡萄相似,与在番茄的表达方式相反.本研究中IAA9在GA3处理后表达上调可能是因为坐果前期,外源GA3促进了内源IAA浓度增加,IAA浓度的升高促使子房和其他部分IAA9的表达量增加,产生大量Aux/IAA蛋白,积累的Aux/IAA蛋白经过泛素化降解,从而调节下游生长素响应基因的表达,引发果实坐果[28].
拟南芥的IAA6蛋白具有完整的4个结构域.水稻中研究发现OsIAA6参与水稻干旱胁迫响应及分蘖生长的调控[29].本研究发现处理前期IAA6表达量较低,清水处理7 d后其他部分IAA6表达量升高,推测可能是不能成功坐果的材料中细胞失水,引起干旱胁迫响应,导致IAA6基因表达量的变化. IAA8是一个典型的Aux/IAA蛋白,拟南芥中,基因IAA8与IAA9的DNA序列同源性很高,其功能缺失的拟南芥突变体可通过改变茉莉酸(JA)的水平而影响花器官的发育[30]. Arase等[31]研究发现,拟南芥中IAA8是一种生长素响应的转录抑制因子,可以与生长素受体TIR1和ARF转录因子相互作用参与侧根的形成.本研究发现子房中外源GA3处理后第3 d开始,IAA8的表达量一直很高,并在21 d达到最高,推测其功能可能与IAA9相似.
大量研究表明生长素信号转导途径中Aux/IAA是与ARF是共同作用的[32-33],如拟南芥中在接受受精信号之前,IAA9与ARF8作为复合物共同抑制果实发育[34].本试验只研究了IAA9和其他5个Aux/IAA基因家族在果实坐果期的表达情况,因此下一步需要克隆更多相关基因,并对关键基因进行转基因功能验证,从而对外源GA3处理促进三倍体枇杷坐果的机理研究提供更多的分子依据.



 下载:
下载: