-
开放科学(资源服务)标志码(OSID):

-
蜡梅Chimonanthus praecox为蜡梅科蜡梅属落叶灌木,是我国特有的传统名贵花木,也是少有的冬季开花观赏植物,具有悠久的栽培历史[1]. 蜡梅因其色如蜜蜡,芳香馥郁而得名,并深受人们喜爱,在我国西南地区被广泛应用于园林绿化[2],也用于香精提取、花茶炮制及窨制. 近年来,高通量测序技术及基因挖掘技术飞速发展,促进了解析基因表达差异、新基因发掘、基因功能研究的进步. 同时,蜡梅cDNA文库[3]和转录组数据库[4-5]的构建,为蜡梅分子生物学研究奠定了基础. 类胡萝卜素是一类含有40个碳且呈黄色、橙红色及红色的类异戊烯聚合物,属于四萜类脂溶性多烯色素,分为胡萝卜素和叶黄素两类[6-8]. 目前,已鉴定出的类胡萝卜素有1 000多种[9],在动物、植物、真菌和细菌中广泛存在. 高等植物的类胡萝卜素主要存在于叶、花、果实、根等有色体的质体中. 存在于叶中的类胡萝卜素在植物光合作用过程中发挥着重要作用[10]. 在果实、根和花等非光合作用组织中,具有不同成分和含量的类胡萝卜素作为特殊代谢产物聚集,不仅使植物的花和果实呈现不同的颜色,还可以分解成有气味和味道的化合物,吸引昆虫传粉和动物传播种子[7, 11-12]. 另外,类胡萝卜素也参与植物激素如脱落酸(abscisic acid,ABA)、独角金内酯(strigolactones)等的生物合成[13-14].
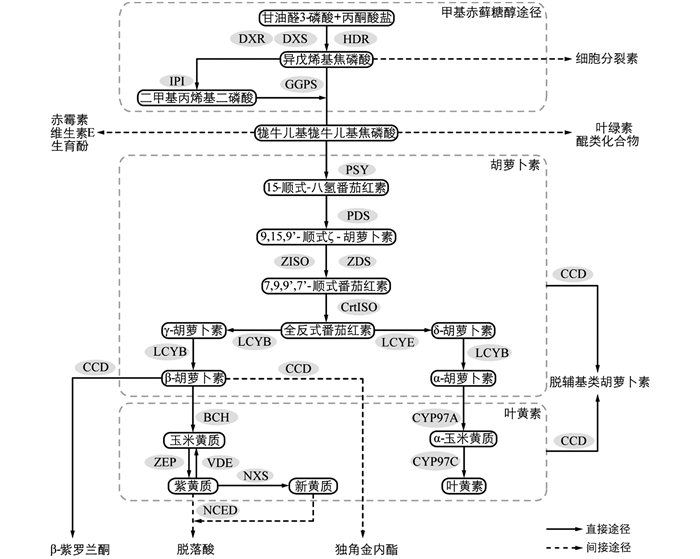
2-C-甲基-D-赤藓糖醇-4-磷酸(2-C-methyl-D-erythritol-4-phosphate,MEP)途径和甲羟戊酸(mevalonate,MVA)途径是植物萜类化合物合成的两个途径[15]. 类胡萝卜素的前体物质异戊烯基焦磷酸(isopentenyl diphosphate,IPP)通过MEP途径合成(图 1),由甘油醛-3-磷酸(glyceraldehyde 3-phosphate,GA3P)和丙酮酸盐(pyruvate)在1-脱氧-D-木酮糖-5-磷酸合酶(1-deoxy-D-xylulose 5-phosphate synthase,DXS)作用下发生缩合反应,生成1-脱氧-D-木酮糖-5-磷酸(1-deoxy-D-xylulose 5-phosphate,DXP),再由1-脱氧-D-木酮糖-5-磷酸还原异构酶(1-deoxy-D-xylulose 5-phosphate reductoisomerase,DXR)催化,经过一系列反应生成IPP[16]. IPP在IPP异构酶(isopentenyl diphosphate isomerase,IPI)的催化下转化为二甲基丙烯基二磷酸(dimethylallyl diphosphate,DMAPP),DMAPP在牻牛儿基牻牛儿基焦磷酸合成酶(geranylgeranyl diphosphate synthase,GGPS/GGPPS)作用下与3个IPP发生缩合反应,生成20碳的牻牛儿基牻牛儿基焦磷酸(geranylgeranyl diphosphate,GGPP). GGPP由八氢番茄红素合成酶(phytoene synthase,PSY)催化,缩合形成15-顺式-八氢番茄红素(15-cis-phytoene). 15-顺式-八氢番茄红素在八氢番茄红素脱氢酶(phytoene desaturase,PDS)和类胡萝卜素异构酶(ζ-carotene isomerase,ZISO)作用下,生成ζ-胡萝卜素. ζ-胡萝卜素在ζ-胡萝卜素脱氢酶(ζ-carotene desaturase,ZDS)和类胡萝卜素异构酶(carotenoid isomerase,CrtISO)作用下形成全反式番茄红素(all trans lycopene). 全反式番茄红素的环化反应由番茄红素环化酶(lycopene β-cyclase,LCYB与lycopene ε-cyclase,LCYE)催化,生成两种不同的类胡萝卜素,即β-胡萝卜素(含2个β环)和α-胡萝卜素(含1个β环和1个ε环)[17]. α-胡萝卜素与细胞色素P450胡萝卜素羟化酶(cytochrome P450 carotene hydroxylase,CYP97)发生反应生成叶黄素(lutein)[18]. β-胡萝卜素与β-环羟化酶(β-carotene hydroxylase,CHY-β/BCH)及玉米黄质环氧酶(zeaxanthin epoxidase,ZEP)反应形成玉米黄质(zeaxanthin)、紫黄质/堇菜黄质(violaxanthin)等物质. 紫黄质可通过新黄质合成酶(neoxanthin synthase,NXS)催化进一步形成新黄质(neoxanthin)[19]. β-胡萝卜素可被类胡萝卜素裂解双加氧酶(carotenoid cleavage dioxygenases,CCD)经过一系列反应裂解,合成独角金内酯[20]、挥发性有机化合物(volatile organic compounds,VOCs)[14, 21]等; 紫黄质和新黄质可被9-顺-环氧类胡萝卜素双氧酶(9-cis-epoxycarotenoid dioxygenases,NCED)催化裂解生成脱落酸合成的前体物质[22].
在植物生长发育过程中,控制类胡萝卜素合成与代谢的机制复杂而多样,不能对单一基因开展研究,需结合多个基因的协同作用进行分析. 本文通过研究类胡萝卜素生物合成与代谢相关基因转录水平的变化及类胡萝卜素含量变化之间存在的相关性,为探索蜡梅类胡萝卜素代谢机制及人工调控蜡梅类胡萝卜素含量提供一定的理论基础,同时也为蜡梅花中黄色可食用色素的提取及茶用蜡梅花的保健功能提供参考.
全文HTML
-
基于西南大学花卉工程技术研究中心构建的蜡梅不同花期转录组数据库[4]及低温打破蜡梅花芽休眠并膨大开放的转录组数据库[5],筛选类胡萝卜素合成与代谢相关的基因. 西南大学校园内长势较一致且无病虫害的蜡梅,采其露瓣期(displayed petal stage,DP)、盛开期(open flower stage,OF)、衰败期(senescing flower stage,SF)的花(图 2),每个样本设置3个生物学重复. 采集到的样本立即用液氮冷冻,然后保存到-80 ℃冰箱中,用于总RNA提取,另取相同的材料用于类胡萝卜素提取.
-
采用有机溶剂法[23]对总类胡萝卜素进行提取与测定. 分别称取露瓣期、盛开期和衰败期蜡梅花0.2 g,液氮研磨后迅速放入10 mL离心管中,加入10 mL 80%丙酮溶液,利用超声波充分振荡直至提取液无色; 吸取上层溶液于新的离心管中,高速冷冻离心机(美国Thermo)5 000 r/min、4 ℃离心10 min,再次吸取上清液,密封避光4 ℃保存备用. 每个处理重复3次. 吸取300 μL上清液于96孔酶标板中,用Varioskan Flash酶标仪(美国Thermo)分别测定其在663 nm,646 nm,470 nm处的吸光值,每个处理重复3次,实验全程避光操作.
-
β-胡萝卜素含量的测定利用高效液相色谱法(high performance liquid chromatography,HPLC). 分别称取2 g露瓣期、盛开期和衰败期的蜡梅花,放入研钵加液氮研磨,然后迅速将粉末置于10 mL离心管中,加入8 mL 80%丙酮溶液,放于4 ℃冰箱避光浸提48 h. 对浸提液低温(4 ℃)离心5 min,用丙酮溶液重复洗脱沉淀并离心,直至沉淀无色,合并上清液,定容至25 mL,过0.22 mm有机滤头,备用,每个处理重复3次; 利用40 ℃旋转蒸发仪蒸干过滤后的浸提液,然后用4 mL乙酸乙酯溶解、测定. 混合标样的制备:以10 mg/L混合标样溶液为标准储备溶液,制备系列质量浓度为0.005,0.01,0.05,0.1,0.25,0.5,0.75,1.0,5.0 mg/L的标准溶液,过0.22 mm有机滤头,备用. 以80%丙酮溶液为空白,利用UV755B紫外可见分光光度计(上海精科),在波长300~700 nm范围内扫描,确定最佳吸收波长,测定类胡萝卜素的总含量; 利用Agilent1100 series高效液相色谱仪(美国Agilent)进行梯度冲洗,测定β-胡萝卜素的含量. 实验全程避光操作,将测得的数据导入GraphPad Prism 9进行方差分析及图形绘制.
-
对蜡梅不同花发育时期转录组数据库中类胡萝卜素相关基因进行表达差异分析,利用TBtools软件[24]进行基因表达差异热图绘制,选取表达差异显著的基因利用生物信息在线分析软件“微生信”(www.bioinformatics.com.cn)进行GO(gene ontology,GO)富集条形图绘制. 对低温打破蜡梅花芽休眠并膨大开放的转录组数据库中类胡萝卜素代谢通路的基因进行表达差异分析,利用TBtools软件进行基因表达差异热图绘制. 所有图片均利用Adobe Illustrator 2020进行组合及美化.
-
选取露瓣期、盛开期、衰败期的蜡梅花为材料,采用北京天根公司生产的RNAprep Pure植物总RNA提取试剂盒进行总RNA提取. 用1% TBE琼脂糖凝胶电泳(北京六一)和NanoDrop 2000c分光光度计(美国Thermo)检测RNA的完整性、纯度及浓度. 以提取的总RNA为模板,用TaKaRa公司生产的PrimeScript RT-PCR Kit反转录试剂盒进行cDNA第一链的合成.
-
选取部分蜡梅类胡萝卜素合成与代谢相关基因,利用实时定量PCR方法进行表达量验证. 选用蜡梅CpActin-b和CpTublin基因作为实时定量PCR的双内参基因. 利用Primer Premier 5.0软件设计内参基因和目标基因的实时定量PCR引物,送华大生物进行引物合成,引物序列见表 1. 应用Bio-Rad IQ5(美国Bio-Rad)进行不同花期各基因实时定量PCR检测,方法同文献[25],将结果采用2-ΔΔCt转化成线性关系,并进行统计学分析.
-
将低温打破蜡梅花芽休眠并膨大开放的转录组数据库[5]中涉及类胡萝卜素生物合成与代谢的基因所编码的蛋白序列上传到STRING v10在线网站(www.string-db.org),进行蛋白相互作用网络构建及相关代谢通路预测,进一步分析直接参与蜡梅类胡萝卜代谢的各蛋白之间的互作关系及作用过程.
1.1. 实验材料
1.2. 蜡梅不同花发育时期类胡萝卜素的提取与含量测定
1.2.1. 总类胡萝卜素的提取与含量测定
1.2.2. β-胡萝卜素提取与含量测定
1.3. 蜡梅不同花发育时期类胡萝卜素合成与代谢基因表达差异分析
1.4. 蜡梅不同花发育时期总RNA的提取及反转录cDNA
1.5. 蜡梅不同花发育时期类胡萝卜素合成与代谢相关基因qRT-PCR分析
1.6. 蜡梅类胡萝卜素合成与代谢蛋白互作网络预测分析
-
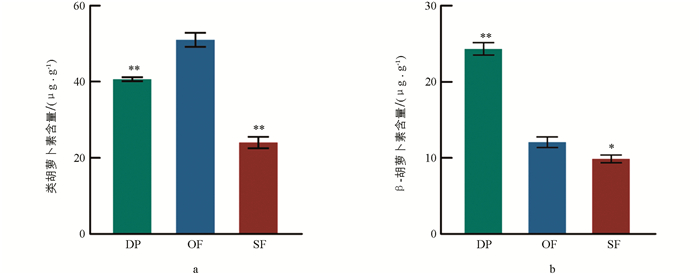
从露瓣期到盛开期,再到衰败期,蜡梅花中类胡萝卜素的总含量呈先升高、后降低的趋势,并在盛开期达到最高,此时含量为51.47 μg/g,分别极显著高于露瓣期和衰败期(图 3a),表明在蜡梅盛开的过程中,类胡萝卜素合成基因,如CpPSY表达量升高. β-胡萝卜素含量整体呈下降趋势,其中露瓣期β-胡萝卜素含量为24.99 μg/g,极显著高于盛开期(近2倍),衰败期与盛开期β-胡萝卜素含量差异显著,但未达到极显著水平(图 3b).
-
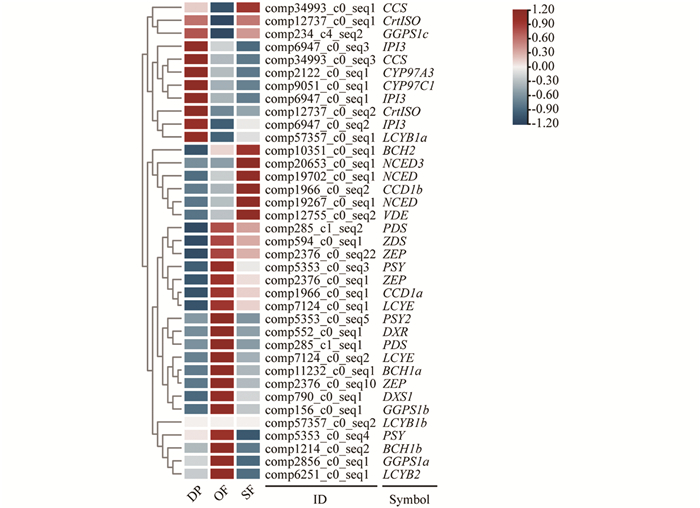
对蜡梅不同花发育时期转录组数据库中与类胡萝卜素合成与代谢相关的基因进行筛选和表达差异分析,利用TBtools软件进行表达差异热图绘制(图 4).
盛开期与露瓣期相比(OF vs DP),CpPSY2表达量上调5.98倍,CpBCH1a,CpBCH2上调均超过8.18倍,表达差异显著; 衰败期与盛开期相比(SF vs OF),CpPSY2表达量下调6.63倍,CpNCED3上调9.53倍,表达差异显著. 从露瓣期到盛开期,再到衰败期,MEP途径中的类胡萝卜素底物合成基因CpDXS1,CpDXR,CpGGPS1a和CpGGPS1b表达量先升高、后降低; 类胡萝卜素合成基因CpPSY,CpPDS,CpZDS,CpLCYE,CpLCYB2,CpBCH1a,CpBCH1b和CpZEP表达趋势同样先升高、后降低; CpCYP97C1和CpCYP97A3表达量持续降低; CpCrtISO,CpGGPS1c和CpIPI3表达量先降低,后略升高; CpLCYB1a和CpLCYB1b几乎不表达,未检测到CpZISO表达; 类胡萝卜素代谢基因CpNCED和CpCCD1b表达量持续升高,CpCCD1a则先升高,后降低; β-胡萝卜素代谢基因CpBCH1a,CpBCH1b在盛开期表达量最高,CpBCH2在衰败期表达量最高.
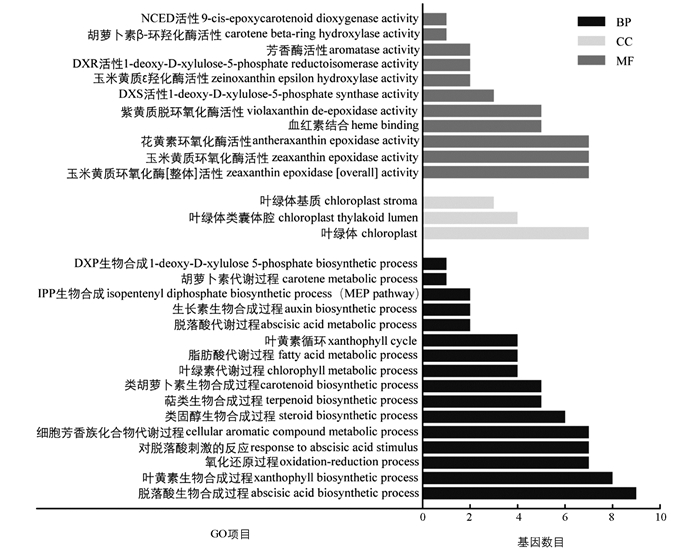
对表达差异显著(log2 Fold Changes≥1或log2 Fold Changes≤-1)的基因进行GO富集分析,筛选直接或间接参与类胡萝卜素合成与代谢的GO Terms作图,结果如图 5所示. GO Terms主要富集在生物过程和分子功能上,其中直接参与类胡萝卜素合成及代谢过程的有类胡萝卜素生物合成(carotenoid biosynthetic process)、萜类生物合成(terpenoid biosynthetic process)、叶黄素生物合成(xanthophyll biosynthetic process)、DXS活性(1-deoxy-D-xylulose 5-phosphate synthase activity)、紫黄质脱环氧化酶活性(violaxanthin de-epoxidase activity)、玉米黄质环氧化酶活性(antheraxanthin epoxidase activity)、叶黄素循环(xanthophyll cycle)及胡萝卜代谢(carotene metabolic)等.
-
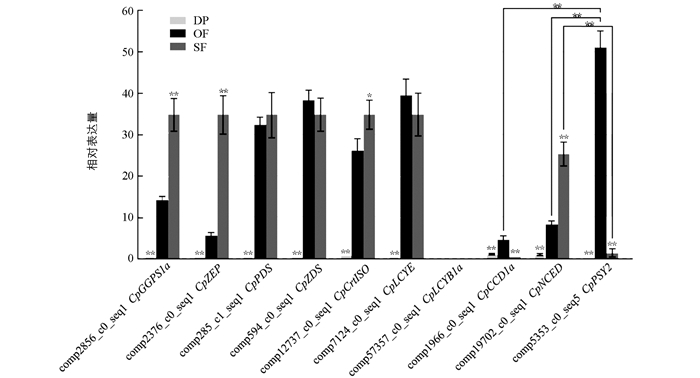
为了明确蜡梅不同花发育时期类胡萝卜素合成与代谢相关基因的表达差异,对蜡梅花露瓣期、盛开期、衰败期类胡萝卜素合成与代谢相关基因的表达特性进行qRT-PCR验证. 结果表明,分析的10个基因在露瓣期表达量均较低,甚至不表达,在盛开期和衰败期表达量较高; 在盛开期,CpPSY2表达量极显著高于CpNCED和CpCCD1a; 在衰败期,CpNCED表达量极显著高于CpPSY2; 盛开期与露瓣期相比(OF vs DP),除未检测到CpLCYB1a表达,其他基因相对表达量均具极显著差异,且盛开期均是露瓣期的4.58倍以上,其中CpPSY2表达量上调高达101.19倍; 衰败期与盛开期相比(SF vs OF),类胡萝卜素合成基因CpPSY2相对表达量下调高达63.10倍,类胡萝卜素代谢基因CpCCD1a下调11.60倍,表达差异极显著(图 6). CpPSY2,CpZDS,CpLCYB1a,CpLCYE,CpCCD1a和CpNCED表达趋势与转录组数据库分析结果基本一致,而另外4个基因表达趋势则与分析结果不完全相同(图 4,图 6); qRT-PCR结果表明CpGGPS1a,CpPDS,CpZEP和CpCrtISO表达量持续上升,在衰败期达到最高(图 6); 而转录组数据分析结果显示CpGGPS1a,CpPDS和CpZEP在盛开期表达量最高,衰败期表达量出现下降,CpCrtISO表达量则在盛开期下降(图 4). 值得注意的是,这些基因表达量的变化不会对类胡萝卜素总含量造成影响,只会影响类胡萝卜素不同组分的含量,比如会使β-胡萝卜素含量降低.
-
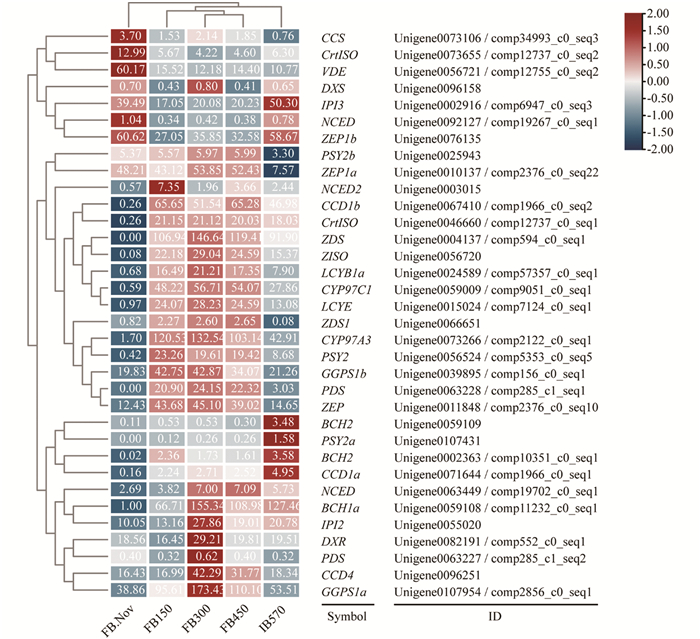
低温对多年生植物的生长发育起着重要作用,在蜡梅花发育过程中只有需冷量(chilling requirement,CR)的积累达到570 CU(chill units,CU)时,蜡梅花蕾(flower bud,FB)才会膨大开放,否则不能开放并逐渐落蕾[5]. 为了进一步研究不同需冷量处理条件下蜡梅花中类胡萝卜素合成与代谢相关基因的变化,基于低温打破蜡梅花芽休眠并膨大开放的转录组数据库,课题组以蜡梅不同需冷量处理(150/300/450 CU)的花蕾(FB150/300/450)、11月份对照花蕾(FB.Nov)及570 CU处理可以始花的花蕾(IB570)样品为材料,筛选并分析各样品中类胡萝卜素合成与代谢相关基因的表达差异. 结果表明,随着需冷量的积累,CpBCH1a,CpBCH2,CpCCD1a和CpCCD1b表达量增加,CpCCD4表达量短暂升高后降低(图 7),使得β-胡萝卜素含量下降; CpLCYB1a和CpLCYE在FB.Nov与IB570中表达量都很低,但在需冷量积累过程中一直保持高表达量; 值得一提的是,类胡萝卜素合成基因CpPSY2在需冷量积累前低表达,开始需冷量积累后表达量升高,到始花期IB570其表达量又出现下降,说明在打破休眠后的始花期之前,类胡萝卜素出现过短暂积累,含量升高; 类胡萝卜素代谢基因CpNCEDs在开始需冷量积累后,表达量也出现了升高,但其表达量低于CpPSY2,因此蜡梅类胡萝卜素含量升高. 样品聚类分析表明,在需冷量积累过程中,FB150/FB300/FB450花蕾中类胡萝卜素合成与代谢基因具有相同的表达模式,而在需冷量积累达到570 CU的始花期花蕾样品(IB570)中,类胡萝卜素合成与代谢基因表达模式会发生变化,以上两种表达模式均不同于低温处理前的对照FB.Nov(图 7).
-
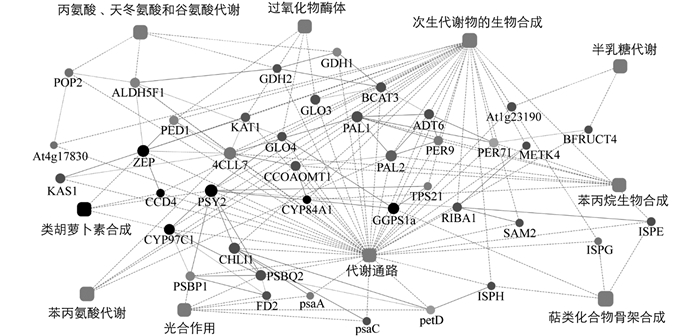
基于前期转录组测序结果[5],利用STRING v10(www.string-db.org)在线软件,以拟南芥Arabidopsis thaliana为参照,对低温诱导开花过程中蜡梅类胡萝卜素合成与代谢相关蛋白进行蛋白相互作用网络预测,明确参与蜡梅类胡萝卜素合成与代谢的蛋白之间的关系及作用通路. 预测结果表明,参与蜡梅类胡萝卜素合成与代谢的蛋白有CpCCD4,CpPSY2,CpZEP和CpCYP97C1(图 8). 同时,CpPSY2与CpGGPS1a有直接作用关系,CpZEP与CpCCD4有直接作用关系. PSY作为类胡萝卜素合成的第一个限速酶,以GGPP为底物合成15-顺式-八氢番茄红素,而GGPP是以IPP和DMAPP为底物,在GGPS的催化下合成的; CCD2和CCD4可裂解玉米黄质,分别生成藏红花素(crocetin/crocinins)和β-柠乌素(β-citraurin)[26-27],而ZEP可环化玉米黄质生成紫黄质. 蛋白互作网络预测结果还表明,CpZEP,CpCCD4,CpCYP97C1,CpPSY2和CpGGPS1a可能是代谢通路的功能蛋白以及次生代谢物生物合成的功能蛋白.
2.1. 蜡梅不同花发育时期类胡萝卜素含量
2.2. 蜡梅不同花发育时期类胡萝卜素合成与代谢基因表达差异
2.3. 蜡梅不同花发育时期类胡萝卜素合成与代谢基因相对表达量
2.4. 类胡萝卜素合成与代谢基因在不同需冷量积累下的表达模式
2.5. 类胡萝卜素相关蛋白互作调控网络预测
-
类胡萝卜素的积累途径包括生物合成、降解和质体中的稳定储存,这些通路都会影响植物组织中类胡萝卜素的最终水平[18]. 在类胡萝卜素的合成通路中,PSY是关键限速酶之一,决定着类胡萝卜素最终的积累水平. 番茄Solanum lycopersicum因其含有丰富的类胡萝卜素而被广泛种植[28-29],转录激活番茄SlPSY1可使其类胡萝卜素生物合成水平显著升高[30],PSY2能够弥补PSY1功能的损失,增加类胡萝卜素的积累[31]. 在蜡梅花发育过程中,从露瓣期到盛开期,CpPSY2大量表达,类胡萝卜素得到积累,含量增加; 在需冷量积累的过程中,CpPSY2表达量也出现了短暂升高,类胡萝卜素含量短暂增加. 增加PSY的作用底物,也可增加类胡萝卜素的积累水平,如在MEP途径过表达DXS以提供合成途径初期的中间产物DXP,进而增加可利用的IPP与DMAPP含量. 在拟南芥中过表达DXS,其类胡萝卜素总积累量达到正常水平的1.5倍[32]. LCYB与LCYE是将番茄红素转化为胡萝卜素的关键酶,LCYB的表达量影响β-胡萝卜素的积累. 研究发现,沉默表达TaLCYB会降低小麦Triticum aestivum籽粒β-胡萝卜素和叶黄素含量,增加番茄红素含量[33]. BCH可环羟化β-胡萝卜素生成玉米黄质,ZEP可环氧化玉米黄质生成紫黄质,紫黄质去环氧化酶(violaxanthin de-epoxidase,VDE)可将紫黄质还原成玉米黄质,它们的相互转化形成了叶黄素循环,这是保护植物免受光损伤的重要机制[34]. BCH的表达量显著影响β-胡萝卜素的积累. 从露瓣期到盛开期,再到衰败期,CpBCH的高表达是蜡梅β-胡萝卜素含量降低的主要原因.
NCEDs和CCDs属于类胡萝卜素裂解加氧酶家族(CCO),是类胡萝卜素代谢的关键酶. 类胡萝卜素通过氧化裂解过程被降解,产生一种被称为脱辅基类胡萝卜素(apocarotenoids)[34]的代谢产物. 脱辅基类胡萝卜素与花、果实的风味及色泽有关,在植物激素产生和防御中也起着重要作用. NCEDs只与特定的类胡萝卜素发生反应,即紫黄质和新黄质,生成脱落酸(ABA)合成的前体物质. 抑制番茄SlNCED1的表达,可以增加番茄红素和β-胡萝卜素的积累[35]. 从蜡梅花蕾开始需冷量积累,经膨大开放,到衰败的整个过程中,CpNCED的表达量持续升高,至衰败期,CpNCED表达量达到最高是蜡梅类胡萝卜素减少的主要原因,其花瓣的黄色也较盛开期有所变浅(图 2); 另外,在人工低温处理(需冷量150 CU,300 CU,450 CU,570 CU)实验中,与11月份花蕾(FB.Nov)相比,随着需冷量的积累,ABA含量逐渐升高,至570 CU打破花蕾休眠(IB570)时达到最高,之后在始花期(IB570)、盛开后期(LB570)和衰败期(WP570)ABA含量逐渐降低[5]; 从露瓣期到衰败期,β-胡萝卜素含量持续降低(图 3b),ABA含量也相应降低[5]; 推测CpNCED表达量的差异可能与ABA含量的变化直接相关. CCD1,CCD2和CCD4的表达可使花呈现多种多样的色彩[36],其中CCD4常有较高的表达水平[37-38],可使β-胡萝卜素的累积明显降低,同时也影响一些植物花和果实的颜色,常使得花、果实呈现白色、淡黄色[39-41]. 在菊花Chrysanthemum morifolium中,CmCCD4的表达被抑制时,白花突变为黄花[42]. CCD1可氧化裂解番茄红素、β-胡萝卜素、叶黄素和玉米黄质等类胡萝卜素,生成含13个碳且具有挥发性的脱辅基类胡萝卜素,如α-紫罗兰酮(α-ionone)、β-紫罗兰酮(β-ionone)、3-OH-β-紫罗兰酮(3-OH-β-ionone)及假紫罗兰酮(pseudoionone)等[43-44],其中β-紫罗兰酮是一种有显著气味和芳香的物质,是植物花香的主要成分之一[45-47]. 玉米Zea mays自交系关联分析中发现ZmCCD1拷贝数越高,类胡萝卜素的含量越低[48]; 在拟南芥、玉米、番茄等植物中还发现CCD1可裂解番茄红素生成挥发性香气物质甲基庚烯酮(6-methyl-5-hepten-2-one,MHO)[49-50]. 在蜡梅花蕾开始需冷量积累,经膨大开放,到盛开的过程中,CpCCD1a和CpCCD1b表达量持续升高,且CpCCD1a在盛开期表达量最高,此时蜡梅香气最为浓郁,可能与CpCCD1a氧化裂解β-胡萝卜素及番茄红素产生芳香物质有关.
-
在蜡梅花发育过程中,从露瓣期到盛开期,再到衰败期,类胡萝卜素总含量先升高、后降低,β-胡萝卜素含量则呈下降趋势. 类胡萝卜素合成基因CpPSY2在盛开期表达量最高,在衰败期表达量下降,同时类胡萝卜素代谢基因CpNCEDs表达量达到最高. 在这3个时期,CpLCYB表达量都不高,CpBCH1和CpBCH2表达量持续增高,这种表达模式不仅限制了β-胡萝卜素含量的增加,也加速了玉米黄质的积累,进而影响类胡萝卜素的组分及含量. 通过调控CpPSY及CpLCYB的表达,改变类胡萝卜素的组分和含量,为人工调控蜡梅花发育不同时期类胡萝卜素的组分及含量提供借鉴和参考.



 下载:
下载: