-
巨魾(Bagarius yarrelli)在中国分布于云南省境内的澜沧江、怒江、元江水系, 其肉呈黄色, 故地方名叫“面瓜鱼”, 隶属于硬骨鱼纲(Osterichthyes), 鯰形目(Siluriformes), 鮡科(Sisordae), 魾属(Bagarius)[1].巨魾是云南特有的大型底栖食肉鱼类, 其体质量可超过50 kg, 体长可超过200 cm.目前对巨魾的研究包括巨魾的生物学特性[2-4]、人工繁殖[5]、疾病防治[6]、胚胎发育[7], 2013年巨魾苗种实现规模化生产, 能够满足增殖放流和养殖生产.
线粒体DNA是目前分子生物学研究的一个重要的领域, 而鱼类mtDNA是鱼类分子系统学研究和群体遗传分析的重要标记[8].近年来线粒体基因组已经被广泛地应用于脊椎动物群体的群体遗传学、系统发生学、保护生物学、比较和进化基因组学、分子进化和系统发育树重建等方面的研究[9-11].本实验所研究的序列为巨魾ND3和ND4L基因片段, 与其他动物一样, 其ND3基因和ND4L基因之间有1个tRNA-Arg基因.韩家波等[12]用PCR产物直接测序法, 测定渤海辽东湾斑海豹15个线粒体的一段从ND3到ND4基因与22个海豹科动物的ND4L基因序列的系统发生分析表明, 海豹科动物分为南半球和北半球两大类群, 斑海豹与港海豹为北半球种类, 亲缘关系最近.陈瑜等[13]对四川黑熊、马来熊、棕熊、美洲黑熊和北极熊的ND3基因和ND4L基因所编码的氨基酸序列进行了多序列比对分析, 结果表明四川黑熊、马来熊、棕熊、美洲黑熊和北极熊的基因序列同源性极高.张四明等[14]采用DNA测序技术首次测定了包括中国特有鲟形目鱼类在内的12种鲟形目鱼类的mtDNA ND4L和ND4基因的序列, 并进行了分子系统学分析, 结果表明环太平洋地区的鲟科鱼类可能有共同的起源.本实验通过巨魾特有的引物序列来克隆其ND3, tRNA-Arg和ND4L基因, 并对其序列多态性进行研究.
HTML
-
本研究的12条巨魾采自云南省河口县, 剪取肌肉组织放于1.5 mL EP管中贴上对应标签, 加入无水乙醇, 置于4 ℃保存备用.
-
高纯度的巨魾鱼基因组DNA的提取采用杜民等[15]的酚-氯仿-异戊醇方法.提取的DNA样品, 用100 mL 1%的琼脂糖凝胶中加入6 μL溴化乙锭(EB)来检测其纯度.利用设计的2对特异性引物Baya9F和Baya9R[15], Baya10F和Baya10R[16], 对提取的DNA进行PCR扩增, 1%的琼脂糖凝胶检测扩增片段, 凝胶成像系统观察并进行拍照保存, 筛选出目的条带利用1.5%的琼脂糖凝胶电泳, 在暗箱式紫外分光仪下切取目的片段, 用琼脂糖凝胶DNA回收试剂盒(天根生化科技北京有限公司)回收DNA, 与PMD18-T载体(北京博迈德生物技术有限公司)进行连接, 利用TOP10感受态细胞进行转化并克隆, 挑选阳性克隆利用M13通用引物进行菌液PCR, 筛选出具有阳性克隆的菌株, 送南京金斯瑞生物科技有限公司测序.
-
利用DNAMAN5.2.2对克隆得到的序列进行手动剪切并与GenBank中巨魾序列进行配对剪切得到目的基因的序列全长, 对序列进行同源性分析; 利用MEGA5.02软件进行碱基组成分析、遗传距离分析并利用最小进化法(Minimum Evolution, ME)构建系统发育树.
1.1. 实验材料
1.2. 实验方法
1.3. 数据分析
-
采用DNAMAN5.2.2和MEGA5.02软件对获得的巨魾序列进行拼接并分析, 表明巨魾鱼ND3基因序列全长为346 bp; 起始密码子为ATG, 终止密码子为T--; 碱基质量分数A为26.2%, T为29.6%, C为28.8%, G为15.4%, 其中A+T质量分数(55.8%)高于G+C质量分数(44.2%), 12个ND3基因序列存在5种单倍型, 发生了5次转换, 1次颠换, 5种单倍型之间平均相对遗传距离为0.007; tRNA-Arg基因序列全长为71 bp, 碱基质量分数A为23.9%, T为23.9%, C为29.6%, G为22.6%, 其中A+T质量分数(47.8%)低于G+C质量分数(52.2%), 12个基因序列完全一致, 只存在一种单倍型; ND4L基因序列全长为297 bp, 起始密码子为ATG, 终止密码子为TAA, 碱基质量分数A为24.6%, T为26.3%, C为33.3%, G为15.8%, 其中A+T质量分数(50.9%)高于G+C质量分数(49.1%), 12个基因序列也完全一致; ND3碱基质量分数见表 1.
-
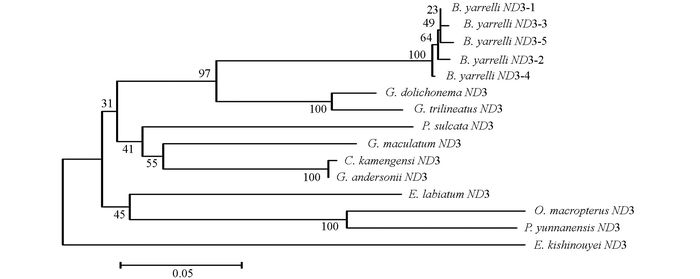
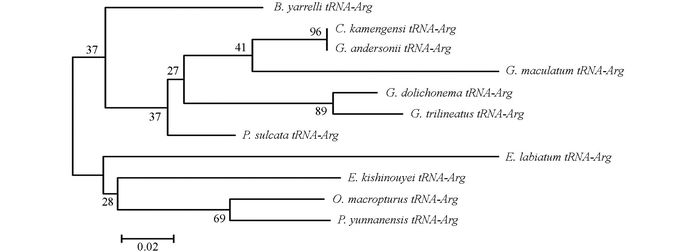
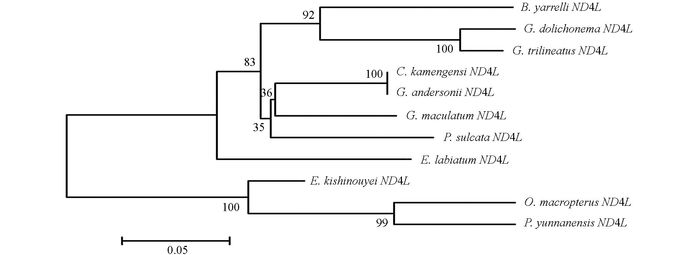
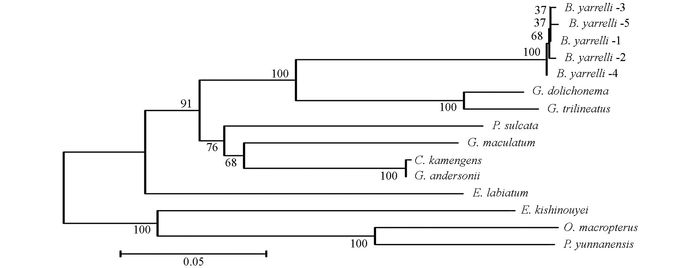
通过DNAMAN5.2.2进行序列比对, 并采用MEGA 5.02软件, 将巨魾鱼与鮡科的其他10种鱼类的ND3, tRNA-Arg和ND4L基因用Minimum Evolution法(ME)构建系统发育树, 系统树各节点的支持率以自展值为1 000次重复抽样检验的百分比结果如图 1, 图 2, 图 3, 图 4.
由母系统树发现巨魾鱼单独聚为一支, 与纹胸鮡属相接近. 10种鮡科鱼类基因片段的种类及在GenBank上面的登录号分别为三线纹胸鮡Glyptothorax trilineatus (NC_021608), 尖须异齿鰋Oreoglanis macropterus (NC_021607), 似黄斑褶鮡Pseudecheneis sulcata (NC_021605), 藏鰋Exostoma labiatum (NC_021603), 拟鰋Pseudexostoma yunnanensis (NC_021604), 凿齿鮡Glaridoglanis andersonii (NC_021600), 贡山鮡Creteuchiloglanis kamengensis (NC_021599), 黑斑原鮡Glyptosternon maculatum (NC_021597), 黄石爬鮡Euchiloglanis kishinouyei (NC_021598), 长丝黑鮡Gagata dolichonema (NC_021596).
2.1. 序列分析
2.2. 分子系统遗传关系
-
鱼类mtDNA基因序列分析中, 13个蛋白质编码基因的结构和功能已了解得比较清楚, 且能用一些通用引物进行扩增, 因而被广泛地用来进行系统进化研究[17].郭宪光[18]基于16S rRNA技术构建鮡科鱼类的分子系统树表明, 鮡科鱼类是一个单系群.王伟等[19]采用RAPD技术在鮡科鱼类属级阶元的分析揭示, 原鮡属与纹胸鮡属亲缘关系较近, 原鮡属是一个比黄颡鱼更为特化的类群, 这与形态学描述的原鮡属为鰋鮡群最原始的属结论相反.周用武等[20]利用Cytb基因片段支持褶鮡属鱼类构成单系群; 而He Shun-ping等[21]分析了鮡科鱼类Cytb基因, 构建了鮡科鱼类系统发育树, 指出鰋鮡鱼类不是一个单系群.但对巨魾的研究还较少, 对巨魾的分类一直都采用传统的形态学分类方法, 而本次实验从分子水平探讨巨魾鱼遗传多样性, 运用PCR扩增和DNA测序技术, 对河口巨魾鱼的线粒体DNA的ND3, tRNA-Arg和ND4L基因序列进行分析, 从而反映出巨魾鱼群体的种质特征, 适用于巨魾鱼群体内的遗传多样性研究.
12条巨魾鱼ND3序列存在5种单倍型, 出现6个变异位点, 5次转换, 1次颠换. 12条巨魾鱼的tRNA-Arg和ND4L基因序列都仅存在1种单倍型, 说明tRNA-Arg和ND4L基因的序列都是高度保守的.从碱基质量分数上分析, 巨魾ND3基因的A+T平均质量分数(55.8%)明显高于G+C平均质量分数(44.2%), 与其他鮡科10个不同ND4L基因序列基本一致; tRNA-Arg基因的A+T平均质量分数(47.8%)略低于G+C平均质量分数(52.2%), 与其他鮡科10个不同种类的tRNA-Arg基因序列组成相反; 巨魾ND4L基因的A+T平均质量分数(50.9%)略高于G+C平均质量分数(49.1%), 与其他鮡科10个不同种类ND4L基因序列的碱基组成基本一致.
线粒体mtDNA ND3, tRNA-Arg和ND4L基因测序表明巨魾个体间的差异极小, 遗传距离从0~0.012, 表明巨魾鱼个体之间无明显差异; 种间的遗传差异极小, 平均差异不到0.3%, 也支持巨魾单独成一支.而将巨魾鱼与其他10种鮡科鱼类的ND3, tRNA-Arg和ND4L基因利用ME法构建的系统进化树, 表明巨魾单独成一支, 与纹胸鮡属相接近, 这一结果与传统的形态学分类基本相似.本实验结果为继续研究其他鮡科鱼类的系统发育提供了有价值的参考资料.





 DownLoad:
DownLoad: