-
开放科学(资源服务)标志码(OSID):

-
甜荞Fagopyrum esculentum和苦荞Fagopyrm tataricum都属于蓼科Polygonaceae荞麦属Fagpyrum的1年生草本作物,起源于中国,具有生长期短、抗逆性强的特点[1-3],富含多种矿质营养元素(铁、钾、锌、硒等)和保健成分(芦丁、槲皮素、不饱和脂肪酸、抗性淀粉、β-葡聚糖等)[4-5],是重要药食兼用作物. 栽培种甜荞花型为异型花(雌雄蕊不等长),分为2种类型,雌蕊长于雄蕊(长柱花,pin)和雌蕊短于雄蕊(短柱花,thrin)[6]. 普通栽培种甜荞自花授粉或同类型花植株之间相互授粉不结实,具有自交不亲和现象[7];同型花雌雄蕊等长,能够自花授粉结实,常见于野生种中. 近年来,国内外多个研究团队利用自交可育野生荞麦F. homotropicum[8],通过种间杂交的方式(F. esculentum和F. homotropicum)培育出自交可育甜荞,即雌雄蕊等长的同型花甜荞[9-12]. Yasui等[13]指出荞麦花的形态是thrum和pin两种形式,通过一个具有两个等位基因状态的基因(S和s),控制thrum(Ss)和pin(ss)形式. 与其他二态型作物一样,花的形态和种内不亲和性均由单个S位点决定,且S-型植株为杂合子(S/s),L-型植株在该位点为纯合隐性(s/s),未发现S/S基因型[13]. 但随后在中国云南省发现了花柱等长自交亲和野生甜荞. Woo等[14]发现自交亲和性受单基因Sh控制,甜荞花型S基因的优势关系为S>Sh>s,自交亲和性等位基因Sh保留了异型性不亲和性,并认为Sh等位基因来源于S超基因复合体[15, 12]. Yasui等[13]在S-Locus基因区注释到32个预测基因,并在这32个预测基因中发现2个与SI相关的候选基因,编码RING/U-box超家族蛋白. Mizuno等[16]利用GBS技术获得甜荞全基因组SNP标记,S等位基因区域内的SNP密度和遗传多样性分析表明该区域具有高度多样性. 由于普通甜荞属于自交不亲和的虫媒传粉作物,花期遇阴雨天气将严重影响结实率,导致每667 m2产量低且十分不稳定(全国平均单产量约为60 kg/667 m2)[11],而通过种间杂交培育的自交可育甜荞,因携带野生荞麦的特性,表现出结实率低、产量低、极易落粒,自交衰退等特点[11, 17]. 因此,甜荞产量与甜荞育性息息相关,对甜荞育性及花柱发育的分子机理进行深入研究具有重要意义. 本研究以自交亲和甜自21和自交不亲和乌克兰大粒荞为材料,分别对甜自21等柱花、乌克兰大粒荞长柱花和短柱花雌雄蕊进行RNA-seq,通过对转录组数据的分析解析甜荞自交不亲和信号传导网络,挖掘自交不亲和相关基因,阐明自交不亲和的分子机理,揭示甜荞育性及花柱发育的分子机制.
HTML
-
2018年秋,自交亲和甜荞品种甜自21和贵甜2号(来源于贵州师范大学),自交不亲和甜荞品种乌克兰大粒荞和酉荞2号分别隔离种植于重庆市北碚区西南大学校内实验基地,常规田间管理,待开花初期将乌克兰大粒荞和酉荞2号长柱花(L-型)植株和短柱花(S-型)植株移栽后隔离种植. 盛花期分别对甜自21和贵甜2号等柱花,乌克兰大粒荞和酉荞2号长柱花和短柱花的花药直径、花冠直径、雄蕊高、花柱高和花柱到花药的距离进行测量并统计分析. 于清晨7:00~9:00点,分别取甜自21和贵甜2号等柱花,乌克兰大粒荞和酉荞2号长柱花和短柱花雌雄蕊,并进行3次生物学重复,样品经液氮速冻后-80 ℃保存,用于RNA提取.
-
采用博日Bioflux的Biospin多糖多酚植物总RNA提取试剂盒提取各个花型雌雄蕊的总RNA. 通过1%琼脂糖凝胶电泳检测总RNA的完整性,使用Agilent Bioanalyzer 2100 System对RNA精确质检. 将检测合格的甜自21等柱花(H),乌克兰大粒荞长柱花(L)和短柱花(S)雌雄蕊总RNA送至北京诺禾致源科技股份有限公司测序,构建cDNA文库,在Illumina HiSeq 4000平台上进行测序. 为了保证信息分析的质量,去除测序得到的原始测序序列(原始数据)里面含有带接头的、低质量的读长后,得到有效数据,然后采用Trinity软件对有效数据进行拼接,得到拼接的转录本. 然后利用Corset层次聚类软件对转录本进行层次聚类,Corset层次聚类后得到最长的Cluster被确定为Unigene,用于后续的分析. 得到的Unigene分别与NCBI(National Center for Biotechnology Information)中的NR和Nt数据库、KO(KEGG ortholog database)数据库、Swiss-Prot蛋白质数据库、Pfam蛋白数据库、GO数据库、KOG/COG数据库进行比对,e-value值设置为小于1e-5. 然后,对注释到GO数据库中的Unigene进行GO功能分类,对注释到COG/KOG数据库中的Unigene进行COG/KOG功能分类分析,对注释到KO数据库中的Unigene进行KEGG代谢途径分析.
-
以Trinity拼接得到的转录组作为参考序列,将每个样品的有效数据在参考序列上做比对,采用RSEM软件,设置bowtie2参数为mismatch 0,对bowtie的比对结果进行统计,得到了每个样品比对到每个基因上的读长数目,并对其进行FPKM转换,进而得到了所有基因的表达水平. 再用DEGseq进行差异分析,筛选阈值为qvalue<0.005且|log2.Fold_change|>1. 然后,分别对差异表达(上调和下调)基因进行GO功能富集分析及KEGG代谢途径富集分析.
-
根据转录组分析结果,筛选出可能与甜荞育性和花柱发育相关的差异表达基因,利用Primer-NCBI在线设计引物. 同时,在自交亲和甜自21和贵甜2号等柱花,自交不亲和乌克兰大粒荞和酉荞2号长柱花和短柱花雌雄蕊总RNA中,利用实时荧光定量PCR进一步验证差异基因的表达特性. 将RNA采用第1链cDNA合成试剂盒(Invitrogen)反转录为cDNA,以cDNA为模板,FeActin为内参,采用TaKaRa(日本)公司的SYBR©Premix Ex TaqTMⅡ(Tli RNaseH Plus)试剂盒,在ABI 7500FAST荧光定量PCR仪(ABI公司,美国)进行荧光定量检测. 每个样品设3次技术重复,依照2-ΔΔCT法计算相对表达量.
1.1. 试验材料
1.2. RNA提取及转录组测序
1.3. 基因表达数据分析
1.4. 实时荧光定量PCR(qRT-PCR)分析
-
选取花柱异长型自交不亲和甜荞品种乌克兰大粒荞(UD)和花柱等长型自交亲和甜荞品种甜自21(TZ)作为实验材料(图 1A). 表型分析结果表明,不同花柱类型花的花冠直径差异不明显,短柱花柱头到花药的距离、花药直径、雄蕊高均大于长柱花,花柱高则是长柱花高于短柱花. 等柱花的花药直径、雄蕊高、花柱高均在长柱花和短柱花之间,且短柱花柱头到花药的距离最小(图 1). 方差分析结果显示,除花药直径外,花冠直径、雄蕊高、花柱高、花柱到花药的距离在长柱花、短柱花和等柱花之间差异均具有统计学意义(p<5%)(图 1).
-
甜自21等柱花(H),乌克兰大粒荞长柱花(L)和短柱花(S)雌雄蕊总RNA样品经转录组Illumina HiSeq 4000测序平台测序后,9个测序样品得到的原始数据,经过严格的质量评估和低质量数据过滤,使其得到高质量的有效数据,Q30(碱基被测错的概率为1‰)均在92%以上,GC含量均在45%左右(表 1),说明得到的有效数据质量和准确度较高,满足后续的数据分析需求.
高质量的有效数据进行拼接后共得到326 245个转录本,平均长度为1 209 bp,N50的长度为1 981 bp. 得到的转录本利用Corset软件进行层次聚类,通过层次聚类后得到Clusters,这些Clusters中具有最长序列的被确定为Unigene,最终得到283 170个平均长度为1 351 bp,N50为2 040 bp的Unigenes.
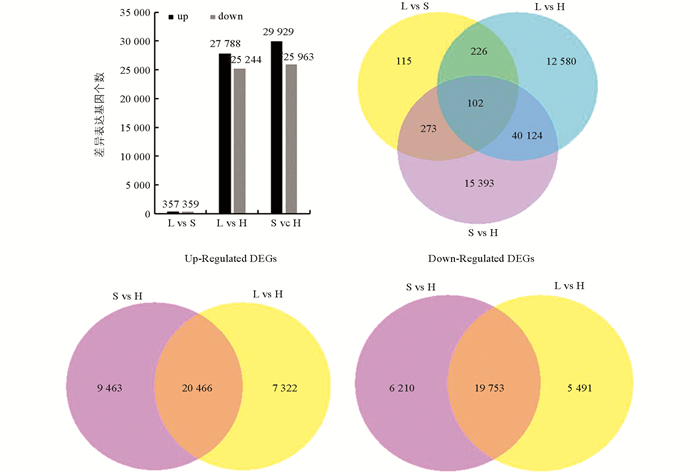
通过分析长柱花(L-型)乌克兰大粒荞、短柱花(S-型)乌克兰大粒荞和等柱花(H-型)甜自21雌雄蕊中的基因差异表达,以等柱花雌雄蕊(H-型)作为对照,在长柱花(L-型)雌雄蕊中上调表达27 788个,下调表达25 244个;在短柱花(S-型)雌雄蕊中上调表达29 929个,下调表达25 963个;长(L-型)、短柱花(S-型)雌雄蕊同时上调基因20 466个,同时下调基因19 753个. 通过对长柱花(L-型)乌克兰大粒荞和短柱花(S-型)乌克兰大粒荞雌雄蕊中的基因进行差异表达分析,共716个差异基因,其中长柱花(L-型)雌雄蕊相对于短柱花(S-型)雌雄蕊上调表达357个,下调表达359个(图 2).
-
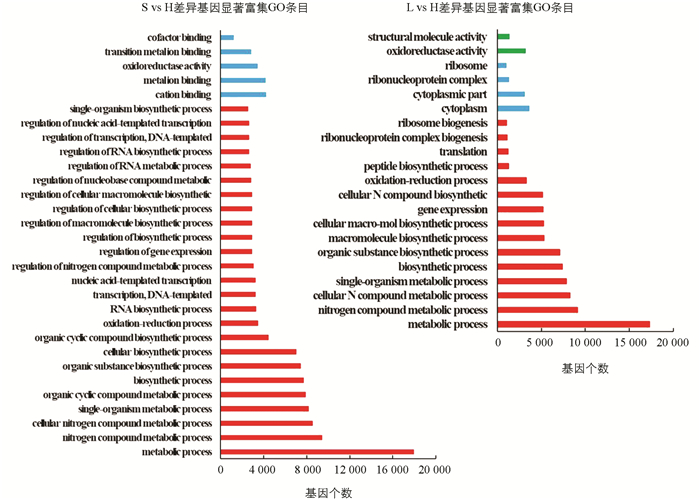
差异表达基因GO富集分析表明,自交不亲和甜荞雌雄蕊(L-型和S-型)与自交亲和甜荞雌雄蕊(H-型)相比较(L vs H,S vs H),差异表达基因特异性富集到氧化还原过程(GO:0055114,oxidation-reduction process)和氧化还原酶活性(GO:0016491,oxidoreductase activity;GO:0016705,oxidoreductase activity,acting on paired donors,with incorporation or reduction of molecular oxygen)最为显著,其次在血红素结合(GO:0020037,heme binding)、铁离子结合(GO:0005506,iron ion binding)、单个有机体代谢过程(GO:0044710,single-organism metabolic process)、转移酶活性(GO:0016758,transferase activity,transferring hexosyl groups)和四吡咯结合(GO:0046906,tetrapyrrole binding)等均有显著富集(图 3),表明这些GO分类中的差异基因与甜荞亲和性有关. 在自交不亲和甜荞中,长柱花(L-型)雌雄蕊与短柱花(S-型)雌雄蕊(L vs S)差异表达基因特异性富集到导管形成过程(GO:0001525,angiogenesis)最为显著,其次为导管发育(GO:0001568,blood vessel development)、导管系统开发(GO:0001944,vasculature development)、导管生成调控(GO:0045765,regulation of angiogenesis)和导管形态发生(GO:0048514,blood vessel morphogenesis)(图 3),表明导管形成相关基因调控甜荞花柱发育.
-
差异表达基因KEGG富集分析表明,自交不亲和甜荞与自交亲和甜荞雌雄蕊(L vs H,S vs H)差异表达基因均显著富集到倍半萜和三萜生物合成途径(ko00909,Sesquiterpenoid and triterpenoid biosynthesis),此外还有花色苷合成途径(ko00942,Anthocyanin biosynthesis),二萜化合物合成过程(ko00904,Diterpenoid biosynthesis),角质、软木脂和蜡质合成(ko00073,Cutin,suberine and wax biosynthesis),β-丙氨酸代谢(ko00410,beta-Alanine metabolism),精氨酸和脯氨酸代谢(ko00330,Arginine and proline metabolism),半胱氨酸和蛋氨酸代谢(ko00270,Cysteine and methionine metabolism)等(表 2),表明萜类化合物可能与甜荞亲和性有关. 在自交不亲和甜荞中,未检测到长柱花和短柱花雌雄蕊(L vs S)差异基因显著富集到某个代谢途径,但植物激素信号转导(ko04075,Plant hormone signal transduction)中的差异基因最多.
-
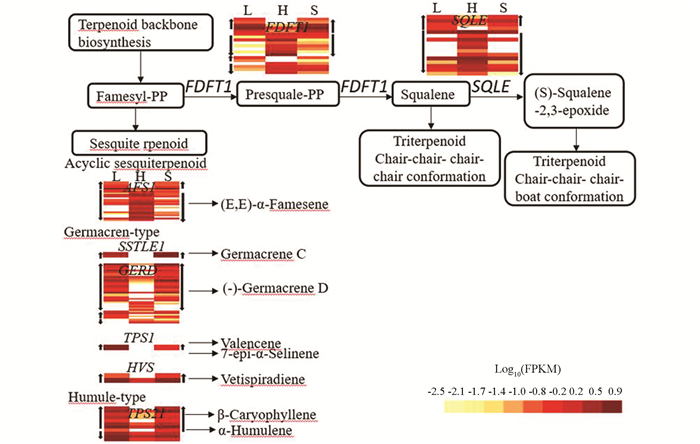
在倍半萜和三萜化合物合成途径(ko00909)中共检测到77个差异基因,与自交亲和甜荞雌雄蕊(H-型)相比,40个DEGs在长柱花雌雄蕊(L-型)和短柱花雌雄蕊(S-型)中同时上调表达,37个DEGs同时下调表达. 其中,三萜化合物(Triterpenoid)合成途径基因FDFT1(farnesyl-diphosphate farnesyltransferase)和SQLE(squalene monooxygenase)以下调表达基因为主. 倍半萜化合物有多个类型,无环型类倍半萜(Acyclic sesquiterpenoid)合成途径基因AFS1(alpha-farnesene synthase)在自交不亲和甜荞长柱花(L)和短柱花(S)雌雄蕊中以下调表达为主;大根香叶烯(Germacren-type)和蛇麻烯(Humule-type)合成途径相关基因SSTLE1(germacrene C synthase),GERD((-)-germacrene D synthase),TPS1(valencene/7-epi-alpha-selinene synthase),HVS(vetispiradiene synthase),TPS21(valencene/7-epi-alpha-selinene synthase)多数为上调表达基因(图 4).
-
植物激素信号转导对于激素触发的生化变化非常重要[16]. 本研究发现,长柱花雌雄蕊相对于等柱花雌雄蕊上调表达基因在“植物激素信号转导”途径(Ko04075)中显著富集. 而且,在Ko04075途径中发现,与自交亲和型等柱花雌雄蕊比较,生长素、茉莉酸和赤霉素信号通路中的差异表达基因在自交不亲和型长、短柱花雌雄蕊中均高表达(图 5). 生长素(AUX/IAA)信号通路中,相对于等柱花(H)雌雄蕊,AUX1,TIR1(transport inhibitor response 1),AUX/IAA,ARF在长柱花(L)和短柱花(S)雌雄蕊中上调表达;茉莉酸相关信号通路与α-亚麻酸代谢密切相关,在茉莉酸信号通路中共发现13个基因,分别为6个GOI1,2个JAR1(jasmonic acid-amino synthetase)和5个MYC2;在赤霉素(GA)信号通路中,与自交亲和型等柱花雌雄蕊比较,自交不亲和型长、短柱花雌雄蕊中表达上调的基因为10个,其中1个GID1(gibberellin receptor GID1),3个DELLA和6个TF.
-
为消除品种特异性影响并验证转录组测序结果的准确性,本研究分别提取自交不亲和的两个甜荞品种乌克兰大粒荞(UD)和酉荞2号(YQ)L-型雌雄蕊、S-型雌雄蕊和自交亲和的两个甜荞品种甜自21(TZ)和贵甜2号(GT) H-型雌雄蕊总RNA,利用qRT-PCR技术,根据已有参考文献对MADS-box[19-20]、MYB[21]及U-box超基因家族[22]等16个差异表达基因进行了qRT-PCR(表 3),检测其在不同品种、不同花柱类型中的表达情况,发现与通过转录组测序FPKM方法计算读长数得出的表达水平,在不同花柱类型表现出一致的表达趋势,表明了转录组测序结果的准确性(图 6).
此外,MADS-box转录因子家族AGAMOUS-like protein (AGL,Cluster-11933.108535)和糖代谢过程中的编码多聚半乳糖醛酸酶基因(Polygalacturonases,PG,Cluster-11933.6467)在短柱花(S-型)雌雄蕊中高表达,长柱花(L-型)雌雄蕊中低表达,等柱花(H-型)中不表达,表明这两个基因可能与甜荞花柱发育有关. 锌指蛋白基因CCCH(Cluster-11933.183409)和U-box超基因家族基因SP11(Cluster-11933.98978)在自交不亲和性甜荞(S-型和L-型)雌雄蕊中表达量较低,甚至不表达,但在自交亲和甜荞(H-型)中均高表达. 相反,转录因子ARF(Cluster-11933.100857,Cluster-11933.100857,Cluster-11933.94328),MYC(Cluster-11933.187734),U-box超基因家族基因SP11(Cluster-11933.123004,Cluster-11933.168503,Cluster-11933.66132,Cluster-11933.98978),转录抑制子SUI1(Cluster-11933.126825)在自交不亲和甜荞雌雄蕊中高表达,在自交亲和甜荞雌雄蕊中低表达,甚至不表达,表明这些基因可能参与调控了甜荞育性.
2.1. 甜荞花表型分析及性状测量
2.2. RNA-seq测序及差异基因筛选
2.3. 差异表达基因GO功能分析
2.4. 差异表达基因KEGG通路分析
2.5. 萜类化合物合成途径基因差异表达分析
2.6. 植物激素相关基因差异表达分析
2.7. 甜荞花柱异型候选基因的qRT-PCR验证
-
甜荞营养价值较高,且耐旱耐瘠薄,是高寒山区重要的经济和药食兼用作物. 甜荞的分子研究起步较晚,研究基础较薄弱. 高通量测序技术的出现,为非模式植物基因组学研究带来了更多的新方法和新方案. 基于高通量测序的从头(donovo)转录组分析可在非模式植物中有效地用于新基因的发现和新分子标记的开发. 目前,已有研究对甜荞籽粒[23-24]、根[25-26]、根和叶[27]、花序[28]、花和子叶[29]进行了转录组测序,并挖掘了相关基因,但有关甜荞自交不亲和性及花柱发育相关的基因鲜有报道. 本研究分别对盛花期自交亲和甜荞品种甜自21等柱花雌雄蕊、自交不亲和甜荞品种乌克兰大粒荞长柱花雌雄蕊和短柱花雌雄蕊进行转录组测序,得到了283,170个Unigenes,其平均长度为1 351 bp,N50为2 040 bp,获得的高质量转录组拼接结果及表达谱数据,将对甜荞育性和花柱发育相关遗传研究提供非常有用的数据资源.
萜类化合物也称类异戊二烯,是由两个异构的5碳骨架(异戊烯基二磷酸(IPP)和二甲基烯丙基二磷酸(DMAPP))组成的最大类别的天然化合物,萜类化合物在自然界中普遍存在,具有各种结构和功能[30]. 萜烯通过参与植物的主要代谢,例如植物激素(脱落酸,细胞分裂素,赤霉素和油菜素甾体)、光合色素(叶绿素和类胡萝卜素)、电子载体(质体醌和泛醌)和内膜系统,对植物的生长和发育至关重要[31-32]. 当植物开花时,一些低分子量的萜类化合物被释放以吸引昆虫进行授粉[33]. 在本研究中,通过对差异基因进行KEGG通路分析发现,自交不亲和甜荞与自交亲和甜荞雌雄蕊(L vs H,S vs H)差异表达基因显著富集到倍半萜和三萜生物合成途径,该途径共检测到79个差异表达基因,42个DEGs在长柱花(L-型)雌雄蕊和短柱花(S-型)雌雄蕊中同时上调表达,37个DEGs同时下调表达,由此推断萜类化合物可能参与调控了甜荞育性发育.
植物激素信号转导对于激素触发的生化变化非常重要[18]. 牵牛花授粉诱导多种植物激素,引起生理和分子变异导致其衰老[34]. 植物生长调节剂生长素基本上参与细胞所有发育过程,包括细胞分裂和扩增,对所有植物发育过程都有影响[35]. 生长素可能直接或间接参与烟草花蕊识别和花粉管伸长[36],在可可[37]、矮牵牛[38]和欧洲橄榄[39]等植物的自交不亲和反应中有重要作用. 在本研究中,植物激素信号转导途径(Ko04075)中的AUX/IAA与色氨酸合成密切相关,该途径中与自交亲和甜荞等柱花雌雄蕊相比,AUX1,TIR1,AUX/IAA,ARF,CH3和SAUR等基因在自交不亲和甜荞长柱花雌雄蕊中均上调表达,推测AUX/IAA相关基因可能参与了甜荞花柱发育.
茉莉酸(JA)在植物发育过程中起着重要的调节作用,如根的生长[40]、雄蕊的发育[41]、毛的形成[42]、开花[43]、叶片的衰老[44]和顶端细胞的形成[45],以及控制对非生物和生物胁迫的不同防御反应. 在拟南芥中,JA诱导R2R3 MYB转录因子MYB21以及MYB24[46],与bHLH转录因子第二亚组(MYC2,MYC3,MYC4和MYC5)[40]来调节后期雄蕊发育[47]. Shi等[48]的研究结果表明,自花授粉梨树花柱中的JA浓度增加,而异花授粉花柱中的JA浓度显著降低. 结合外源JA处理,Shi等[48]认为S-RNase的表达水平降低与JA信号级联有关. 本研究中α-亚麻酸代谢途径在茉莉酸信号作用下,MYC2在自交不亲和甜荞长、短柱花雌雄蕊中上调表达,自交亲和甜荞、雌雄蕊中下调表达. 结果表明JA信号对bHLH转录因子MYC2有影响,从而参与调控甜荞自交亲和性.
赤霉素(GA)是植物中许多发育过程必不可少的植物激素,包括种子发芽、茎伸长、叶片膨胀、毛状体发育、花粉成熟和开花诱导[49]. 植物中存在的具有生物活性的GA较少,主要包括GA1,GA3,GA4和GA7,是碱性二萜类羧酸骨架的衍生物[50]. DELLA是GA反应的关键细胞内阻遏物,抑制种子发芽、生长和几乎所有已知的GA依赖性过程,而GA则减轻其抑制活性[49]. DELLA是植物特异性GRAS家族中假定的转录调节因子的子集,拟南芥中有5个DELLA,它们在抑制GA反应中起着独特或重叠的功能[51-52],其中RGA和GAI抑制营养生长和花诱导[53],RGA,RGL1和RGL2共同调节花的发育[52]. 本研究的Ko04075途径,在GA作用下二萜化合物生物合成通路中,与自交亲和甜荞相比TF,DELLA和GID1基因在自交不亲和甜荞长、短柱花雌雄蕊中均上调表达. 因此,本研究推测赤霉素通过影响萜类化合物的合成,进而参与调控甜荞自交不亲和性.
-
本研究获得了自交不亲和甜荞长、短柱花和自交亲和甜荞等柱花雌雄蕊的基因表达谱数据,共获得283 170条Unigenes,其中有68 813个差异表达基因(DEGs). 自交不亲和甜荞中长、短柱花雌雄蕊中的差异基因主要富集在导管形成和导管发育过程中;自交不亲和甜荞(S-型和L-型)与自交亲和甜荞(H-型)雌雄蕊中差异基因主要富集在倍半萜和三萜化合物合成途径中;自交不亲和甜荞长、短柱花雌雄蕊中差异基因(L vs S)主要富集在植物激素信号转导途径中,生长素、茉莉酸和赤霉素信号通路中的差异表达基因在自交不亲和型长、短柱花雌雄蕊中均高表达;ARF,MYC,MADS和U-box超基因家族中某些基因参与了甜荞育性发育.








 DownLoad:
DownLoad: