-
斑马鱼Danio rerio是一种小型热带淡水鱼类,因其独特的优势,如饲养成本低,胚胎半透明,体外受精,个体发育快,子代产量高等,近年来已经发展成为重要的脊椎模式动物[1].目前,多种遗传技术可以有效地应用在斑马鱼中,如大规模正向遗传筛选[1]、Tol2转座子系统构建转基因鱼[2],Morpholino注射实现基因敲降[3],ZFN(Zinc-finger nucleases)、TALEN(transcription activator-like (TAL) effector nucleases)、CRISPR(Clustered regularly interspaced short palindromic repeats)/Cas9等基因编辑技术进行靶向基因敲除[4-6]等.
CRISPR系统首先在细菌和古细菌中被发现,是其用来保护自身不受外源DNA侵害的一种防御系统[7].基于CRISPR系统工作原理开发的CRISPR/Cas9技术不仅被广泛应用于诸多物种的单基因[8-18]或多基因敲除[19],而且该技术还可以实现DNA片段在基因组中定点插入[20],同时还有报道显示,改造过的CRISPR系统可以对沉默的基因再激活[21]. CRISPR/Cas9毫无疑问地成为了当前最流行且最有应用前景的基因编辑技术.
斑马鱼的成年肾脏是由胚胎期的前肾发育至中肾而形成,相较于哺乳动物的后肾,其肾单位数量较少,仅有数百个肾单位[22].斑马鱼胚胎期的前肾则更加简单,仅由2个肾单位(2个前肾管和融合在一起的2个肾小球)组成,虽然在形态方面它与哺乳动物的肾单位存在较大差异,但是其解剖结构和分子标记表明斑马鱼前肾与哺乳动物肾单位类似[23-24].器官的发生必然受到严格的基因调控,对这一过程的了解将有利于我们更好地理解这些器官的发育过程,并且为研究相关疾病的分子机制以及寻找新的治疗方法提供理论基础.胚胎期基因的表达空间位置暗示它可能在此发挥功能,为研究斑马鱼前肾的发育,本实验选取了10个在斑马鱼前肾表达的基因atp1a1a1,atp1a1a.2,atp1a1a.3,atp1a1a.4,atp1a1a.5,pkd2,aqp3a,bmper,isl2a和tbx2a,采用CRISPR/Cas9技术对它们进行敲除,经过三代的筛选,我们获得了这10个基因的稳定遗传突变体,并且对纯合突变体进行了初步的表型分析,其中2个突变体表现出早期胚胎发育缺陷.本实验的研究结果表明,CRISPR/Cas9技术在斑马鱼中可以快速高效地进行基因敲除.本实验产生的突变体为后续斑马鱼前肾发育的深入研究提供了重要的材料.
HTML
-
野生型斑马鱼品系AB Tüebingen用于本研究.斑马鱼的饲养和繁殖均参照实验动物管理委员会条例规定的标准进行.
-
pXT7-hCAS9质粒用于制备Cas9 mRNA;pMD19T-gRNA质粒用于制备体外转录gRNA所需的DNA模板(北京大学熊敬维教授、张博教授提供).
-
atp1a1a1,atp1a1a.2,atp1a1a.3,atp1a1a.4,atp1a1a.5,pkd2,aqp3a,bmper,isl2a和tbx2a gRNA正向引物由上海Invitrogen公司合成,基因gRNA靶位点附近基因组DNA片段扩增引物由上海生工生物技术公司合成,引物序列见表 1.
-
AxyPrepTM plasmid Miniprep Kit和AxyPrepTM PCR cleanup Kit购自Axygen公司;T7 RNA polymerase购自Invitrogen公司;体外转录试剂购自Ambion公司.
-
交配前一日,将1对雌、雄成年斑马鱼放入交配盒中,用挡板隔开,过夜.次日早晨移走隔板,让2条鱼交配产卵,30 min后收集受精卵,在0.03%的海盐水中于28.5 ℃恒温培养箱饲养.
-
gRNA靶位点一般是一个小于20个碱基的DNA片段,可根据靶位点序列要求进行人工选择[必需条件:紧邻靶位点3'端的3个碱基为PAM(protospacer adjacent motif)区,其序列必需为NGG;次要条件:本实验选用pMD19T-gRNA质粒用于gRNA合成,此质粒采用T7启动子,为提高转录效率,靶位点的5'端最好为GGG/A,若靶位点5'序列无法满足此要求,可在合成gRNA正向引物中的靶位点前加入此序列],也可以通过http://zifit.partners.org/ZiFiT/CSquare9Nuclease.aspx网站进行gRNA靶位点预测和选择.靶位点可以在正义链或者反义链上.将选择好的靶位点序列在Ensembl网站与既有斑马鱼基因组序列进行比对,确认其唯一性.由于斑马鱼个体间存在较多单核苷酸多样性,需对待用野生型斑马鱼的靶位点序列进行检测.拔取野生型待用成鱼2~3片鱼鳞,放入加有10 μL 50 mmol/L NaOH溶液的PCR管中,95 ℃加热10 min,冷却后加入1 μL 1 mol/L Tris 8.0溶液,混合均匀,获得基因组DNA,以此为模板用表 1中的引物PCR扩增靶位点附近基因组DNA片段,并对PCR产物测序,确认待用成鱼基因组中的靶位点序列与选择的是否一致,若不一致则重新选择靶位点;若靶位点序列为杂合子,则重新选择待用成鱼.
-
pMD19T-gRNA质粒中含有体外转录合成gRNA的必要元件,如T7启动子等,gRNA正向引物包含有选择好的靶位点序列,配以通用的gRNA反向引物,以pMD19T-gRNA质粒为模板,PCR扩增得到DNA,并以此PCR产物为模板体外转录合成gRNA.
-
将体外转录合成的Cas9 mRNA和gRNA混合(Cas9 mRNA终质量浓度300 ng/μL、gRNA终质量浓度50 ng/μL),显微注射入1-cell期的斑马鱼受精卵细胞中,每个胚胎注射1 nl,注射过的受精卵置于28.5 ℃恒温培养箱培养.在1 dpf (day post fertilization),随机吸取10枚注射后的胚胎和10枚未注射的胚胎,分别放入EP管中,加入100 μL 50 mmol/L NaOH溶液,95 ℃加热裂解10 min,冷却后加入10 μL 1 mol/L Tris 8.0溶液混合均匀,得到基因组DNA,以此为模板进行PCR扩增靶位点附近基因组DNA片段,并将PCR产物测序.如果gRNA工作,相较于未注射的对照组,注射组的测序峰图会在靶位点序列附近呈现出低水平套峰.
-
将Cas9 mRNA和工作的gRNA共注射的胚胎饲养至成鱼,即为F0代.将成鱼F0与野生型外交产卵,在1 dpf,随机吸取10枚胚胎,用1.2.3中的方法提取基因组DNA、PCR扩增和产物测序,检测靶位点序列附近是否呈现出套峰,用来确定F0子代是否携带突变,若携带突变,将此F0与野生型成鱼交配,产下的胚胎饲养至成鱼,即为F1代.拔取2~3片F1成鱼鱼鳞,裂解获得基因组DNA,进行PCR扩增、产物测序以确定F1个体是否为杂合子及基因突变类型.将携带移码突变的F1杂合子与野生型成鱼外交繁殖F2代.在F2成鱼中拔取鳞片,提取基因组DNA,通过PCR产物测序筛选F2杂合子.
1.1. 材料
1.1.1. 实验动物
1.1.2. 质粒
1.1.3. 引物
1.1.4. 主要试剂
1.2. 方法
1.2.1. 胚胎的收集
1.2.2. 基因gRNA靶位点的选择
1.2.3. gRNA的合成
1.2.4. CRISPR/Cas9活性检测
1.2.5. 基于CRISPR/Cas9技术的斑马鱼突变体构建
-
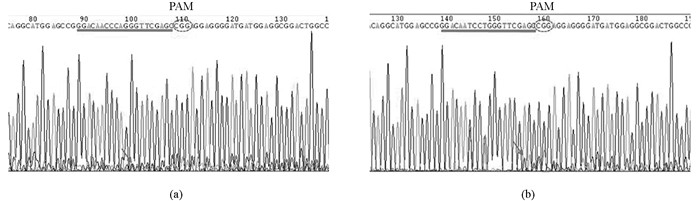
在选取的10个斑马鱼前肾表达基因中,每个基因至少获得了1条工作的gRNA.以pkd2基因靶位点测序峰图为例,如图 1a所示(箭头指示套峰出现的位置,直线标出的是gRNA靶位点,虚线椭圆为PAM区).本文中所用gRNA对应的Cas9靶位点序列见表 2.
-
在F0筛选中,每个拟敲除基因至少获得了一个子代携带突变的F0成鱼.以pkd2基因靶位点测序峰图为例,如图 1b所示.当F1胚胎所含突变类型较少时,可直接通过读取套峰图来确定F1子代的突变类型.
-
F1中含有野生型和杂合子,通过筛选最终获得了这10个斑马鱼前肾表达基因共15个移码突变,基因突变型如表 2所示.
-
F2杂合子成鱼自交后收取胚胎,在显微镜下连续观察至5.5 dpf,以确定在早期发育过程中这些基因突变是否会导致斑马鱼胚胎产生发育缺陷.实验结果显示,在这10个突变基因中,我们只发现了2个基因atp1a1a.2和pkd2突变后可导致斑马鱼胚胎早期发育缺陷,如图 2c,图 3c所示,而其他8个基因突变后在胚胎发育早期,至少5.5 dpf之前我们观察不到明显的胚胎发育异常.
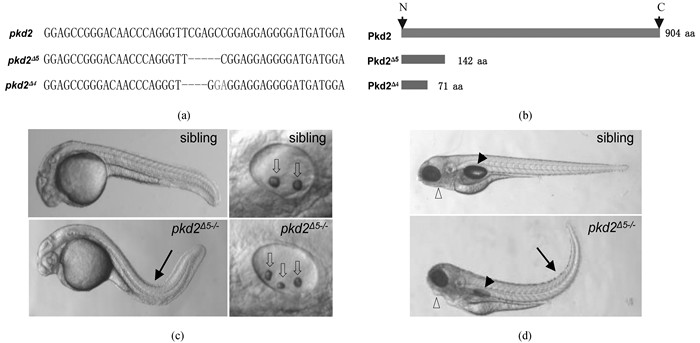
斑马鱼pkd2基因编码多囊蛋白2,与人类PKD2基因同源. PKD2是人类常染色体显性多囊肾病(Autosomal dominant Polycystic Kidney Disease,ADPKD)的主要致病基因[25].多囊蛋白1与多囊蛋白2共同维持正常肾小管形态发生和分化.当PKD1或PKD2发生突变,可导致肾小管细胞内信号转导异常,使得正常肾小管形态不能维持,发生肾囊肿[26].笔者在斑马鱼中运用CRISPR/Cas9技术对pkd2进行靶向敲除,经过三代筛选获得了2种pkd2基因的突变类型Δ5和Δ4,如图 2a所示.这2种pkd2基因突变都造成了阅读框的改变并分别引入一个提前终止密码子,理论上突变基因只能编码截断蛋白,pkd2Δ5基因突变导致产生1个有142个氨基酸的蛋白,而pkd2Δ4突变mRNA则只编码了71个氨基酸,蛋白长度示意图见图 2b.斑马鱼pkd2基因突变纯合子是隐性胚胎期致死的,但它并不像人类同源基因突变一样,产生肾囊肿.在1 dpf,pkd2Δ5-/-突变体鱼体躯干发生向背部的弯曲(图 2c,箭头所示),同时突变体的耳朵结构也异常,出现多个较小的耳石(图 2c,空心箭头所示);当突变体发育至4 dpf,躯干弯曲的表型仍明显(图 2d,箭头所示),并进一步表现出异常的腮弓及未能正常充气的鱼鳔(图 2d,分别由空心三角和黑色三角所示).突变体pkd2Δ4-/-的表型与pkd2Δ5-/-一致.上述表型与已发表的由逆转录病毒插入pkd2基因产生突变体的结果一致[27],表明本实验的基因敲除是可信的.
斑马鱼的ATPase,Na+/K+ transporting,alpha 1a.2 polypeptide (Atp1a1a.2) 是一种具有ATPase酶活性的Na+/K+离子通道膜蛋白,它的人同源基因与醛固酮腺瘤和继发性高血压疾病相关[28].同样,笔者用CRISPR/Cas9技术对atp1a1a.2进行基因突变,最终获得了一种atp1a1a.2基因突变类型Δ1+15,如图 3a所示.此突变造成atp1a1a.2阅读框的改变.同时也引入了一个提前终止密码子,导致突变基因只编码145个氨基酸,见图 3b.斑马鱼atp1a1a.2基因突变纯合子也是隐性胚胎期致死的,它在胚胎发育早期(3 dpf前)正常,随后才显示出异常.在5.5 dpf,atp1a1a.2Δ1+15-/-突变体外观基本正常,但耳石偏小(图 3c,空心箭头)、腹侧鱼鳍皱缩及部分缺失(图 3c,虚线椭圆圈)、腹部存在多个块状的细胞团(图 3c,黑色星状).上述表型中内耳缺陷与文献中Morpholino敲降atp1a1a.2基因表达的结果一致[29],但是腹侧鱼鳍的表型则未见报道.
2.1. CRISPR/Cas9 gRNA活性检测
2.2. 子代携带突变的F0筛选
2.3. F1杂合子的筛选
2.4. 斑马鱼前肾表达基因突变纯合子的表型鉴定
-
笔者通过搜索Zfin网站上具有斑马鱼前肾表达的基因,筛选出其中10个atp1a1a1,atp1a1a.2,atp1a1a.3,atp1a1a.4,atp1a1a.5,pkd2,aqp3a,bmper,isl2a和tbx2a,并运用目前最热门的CRISPR/Cas9基因编辑技术对这些基因进行敲除.经过三代筛选,这10个基因均已获得至少一种可稳定遗传的移码突变,结果表明CRISPR/Cas9技术在斑马鱼中进行基因敲除是快速高效的.此外,笔者初步分析了纯合突变体早期胚胎发育的表型.在10个基因的突变体中,2个基因atp1a1a.2和pkd2突变后可以导致斑马鱼胚胎早期发育缺陷,并且是隐性致死的,其表型与已发表的突变体或Morpholino敲降基因表达的结果一致;其他8个基因突变后,至少5.5 dpf之前,我们观察不到明显的胚胎发育异常.本研究结果表明,大部分斑马鱼基因突变并不导致早期胚胎发育缺陷,这与世界上许多实验室的结果一致.导致这一现象原因有多种:① 在时期上,我们只观察到5.5 dpf,可能是这些基因在胚胎发育早期的确不是必需的,但是有可能在后续阶段发挥功能. ② 纯合突变体是由杂合子自交产生的,母源效应的作用导致这些突变体不产生早期表型;③ 未能敲除所有异构体.一般靶位点选取在基因阅读框中近5'端,若这一基因有多个异构体,这种靶位点的人为选择有可能只敲除了该基因的一种异构体,而在机体里还有其他异构体存在,其他异构体有可能在功能上进行了补偿;④ 基因敲除后,机体存在补偿机制.有研究发现基因敲除后,由其他基因进行了功能补偿,造成突变体没有表型[30].






 DownLoad:
DownLoad: